y veg effort
1 15 Veg1 5
2 18 Veg1 5
3 12 Veg2 5
4 14 Veg2 5
5 12 Veg2 5
6 14 Veg2 5
Veg1 Veg2 Veg3 Veg4 Veg5
2 20 20 10 10 Representing variation among individuals arising, for example, from genetics, location, or experience.
Studying phenomena operating at more than one spatial scale or level of ecological organization.
Modeling a process as well as uncertainty that results from imperfect observations of the process.
Understanding changes in states of ecological systems that cannot be observed directly. These states arise from “hidden” processes.
Random Effect / Multi-level Model
latent/unobserved state or process
“People are always asking me if I want to use a fixed or random effects model for this or that.”
“I always reply that these terms have no agreed-upon definition.”
“People with their own favorite definitions don’t always realize that other definitions are out there. Worse, people conflate different definitions”.
repeated measurements of individual within population
repeated measurements at a small spatial scale that is part of a larger one
Study goals
Estimate state-wide average mass
Understand site-level variation
We sample Amur Leopards in five different vegetation types of Land of the Leopard National Park.
Interested in detection rate by vegetation type and overall.
We sample Amur Leopards in five different vegetation types of Land of the Leopard National Park.
Detection Rate = Independent Counts / Effort
y veg effort
1 15 Veg1 5
2 18 Veg1 5
3 12 Veg2 5
4 14 Veg2 5
5 12 Veg2 5
6 14 Veg2 5
Veg1 Veg2 Veg3 Veg4 Veg5
2 20 20 10 10 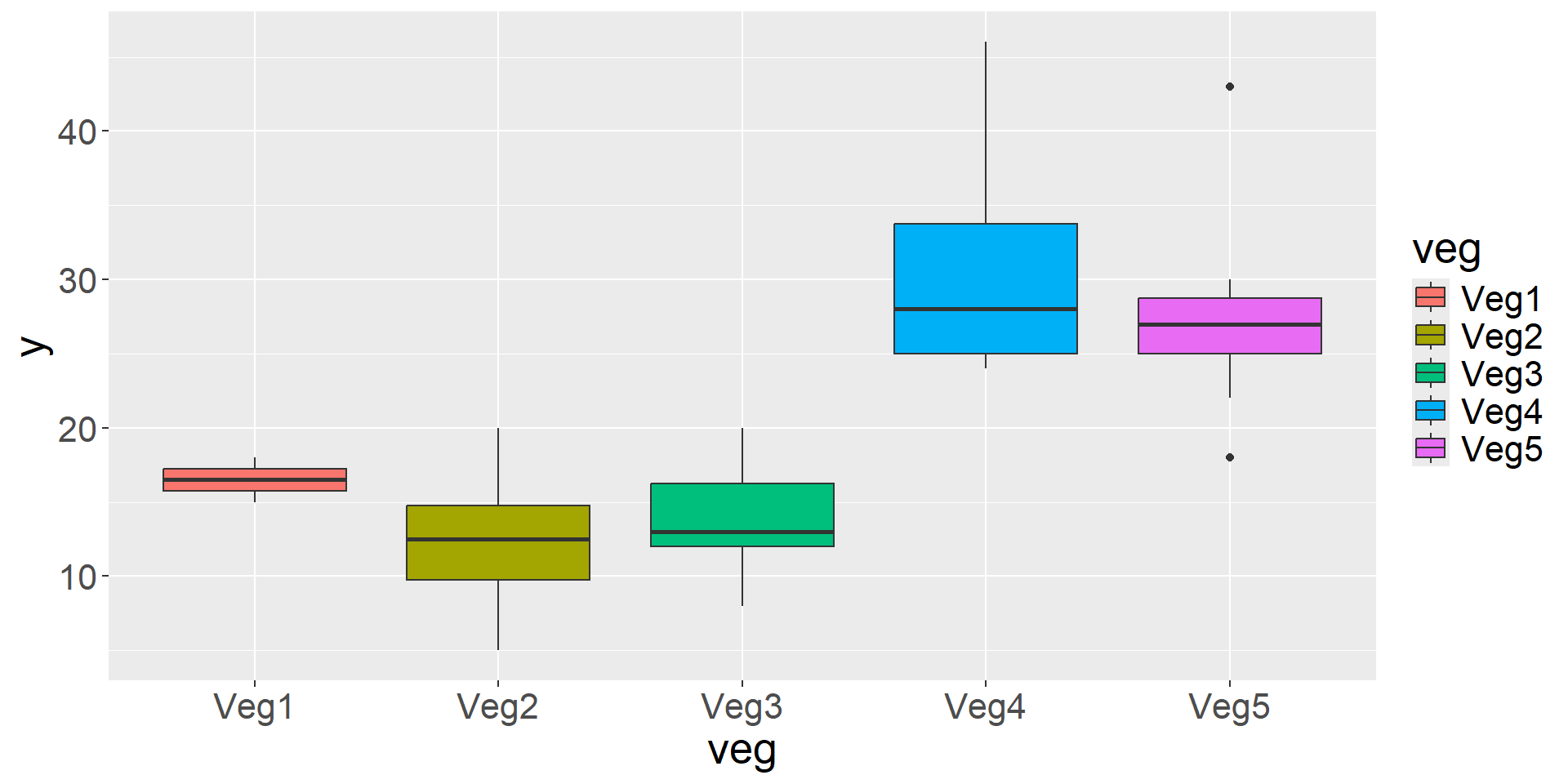
Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
--- 52 rows omitted. See ?avg_predictions and ?print.marginaleffects ---
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
18.4 0.545 33.8 <0.001 827.7 17.3 19.5
Columns: rowid, estimate, std.error, statistic, p.value, s.value, conf.low, conf.high, y
Type: response Family: poisson ( log )
Formula: y ~ veg
Data: dat
AIC BIC logLik deviance df.resid
369.4 380.1 -179.7 359.4 57
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.93764 0.04235 69.37 < 2e-16 ***
veg1 -0.13427 0.14133 -0.95 0.342
veg2 -0.42398 0.06498 -6.52 6.82e-11 ***
veg3 -0.29146 0.06262 -4.65 3.25e-06 ***
veg4 0.47681 0.06138 7.77 7.94e-15 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1marginaleffects::predictions(model2,
newdata = data.frame(veg=c("Veg1","Veg2","Veg3","Veg4","Veg5")),
re.form=NA)
Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
16.5 2.872 5.74 <0.001 26.7 10.9 22.1
12.4 0.786 15.72 <0.001 182.5 10.8 13.9
14.1 0.840 16.79 <0.001 207.8 12.5 15.7
30.4 1.744 17.44 <0.001 223.7 27.0 33.8
27.4 1.655 16.55 <0.001 202.0 24.2 30.6
Columns: rowid, estimate, std.error, statistic, p.value, s.value, conf.low, conf.high, veg, y
Type: response Family: poisson ( log )
Formula: y ~ 1 + (1 | veg)
Data: dat
AIC BIC logLik deviance df.resid
384.4 388.6 -190.2 380.4 60
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
veg (Intercept) 0.1299 0.3604
Number of obs: 62, groups: veg, 5
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.9391 0.1662 17.69 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1$veg
(Intercept)
Veg1 -0.1103110
Veg2 -0.4126484
Veg3 -0.2851561
Veg4 0.4635429
Veg5 0.3612502# A tibble: 5 × 9
effect component group level term estimate std.error conf.low conf.high
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 ran_vals cond veg Veg1 (Interce… -0.110 0.206 -0.514 0.294
2 ran_vals cond veg Veg2 (Interce… -0.413 0.173 -0.751 -0.0738
3 ran_vals cond veg Veg3 (Interce… -0.285 0.172 -0.622 0.0520
4 ran_vals cond veg Veg4 (Interce… 0.464 0.172 0.126 0.801
5 ran_vals cond veg Veg5 (Interce… 0.361 0.173 0.0230 0.699 #Predictions - does not include RE uncertainty
preds = predict(model3,
newdata=data.frame(veg=c("Veg1","Veg2","Veg3","Veg4","Veg5")),
type="link",
re.form=NULL,
se.fit = TRUE
)
preds$LCL = exp(preds$fit-1.96*preds$se.fit)
preds$UCL = exp(preds$fit+1.96*preds$se.fit)
preds$fit = exp(preds$fit)
data.frame(preds) fit se.fit LCL UCL
1 16.92475 0.15877885 12.39843 23.10348
2 12.50889 0.06300056 11.05583 14.15291
3 14.20980 0.05890828 12.66030 15.94893
4 30.04303 0.05761047 26.83519 33.63433
5 27.12181 0.06039730 24.09392 30.53021\[\begin{align*} y_{i} \sim& \text{Poisson}(\lambda_{i})\\ \text{log}(\lambda_{i}) =& \mu + \alpha_{j[i]}\\ \alpha_{j} \sim& \text{Normal}(0, \sigma^2_{\alpha}) \end{align*}\]
Leopard detection varies by cover and veg, where the effect of cover comes from a shared distribution
y cov veg
1 3 0.40161608 Veg1
2 0 -0.12360373 Veg1
3 3 0.62883136 Veg1
4 0 0.07607150 Veg1
5 2 0.04150764 Veg1
6 0 -0.39289231 Veg1Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: poisson ( log )
Formula: y ~ cov + (cov | veg)
Data: dat2
AIC BIC logLik deviance df.resid
14274.7 14295.8 -7132.4 14264.7 495
Scaled residuals:
Min 1Q Median 3Q Max
-4.798 -2.443 -1.630 -0.946 145.188
Random effects:
Groups Name Variance Std.Dev. Corr
veg (Intercept) 0.4896 0.6997
cov 0.1392 0.3732 0.14
Number of obs: 500, groups: veg, 5
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.6210 0.3137 5.166 2.39e-07 ***
cov 0.1121 0.1683 0.666 0.505
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
cov 0.140 \[ \begin{aligned} \operatorname{y}_{i} &\sim \operatorname{Poisson}(\lambda_i) \\ \log(\lambda_i) &=\alpha_{j[i]} + \beta_{1j[i]}(\operatorname{cov}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\mu_{\alpha_{j}} \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for veg j = 1,} \dots \text{,J} \end{aligned} \]
model specifications
predictions with uncertainty
Testing RE’s
Shrinkage/Regularization
When to use a random effect?