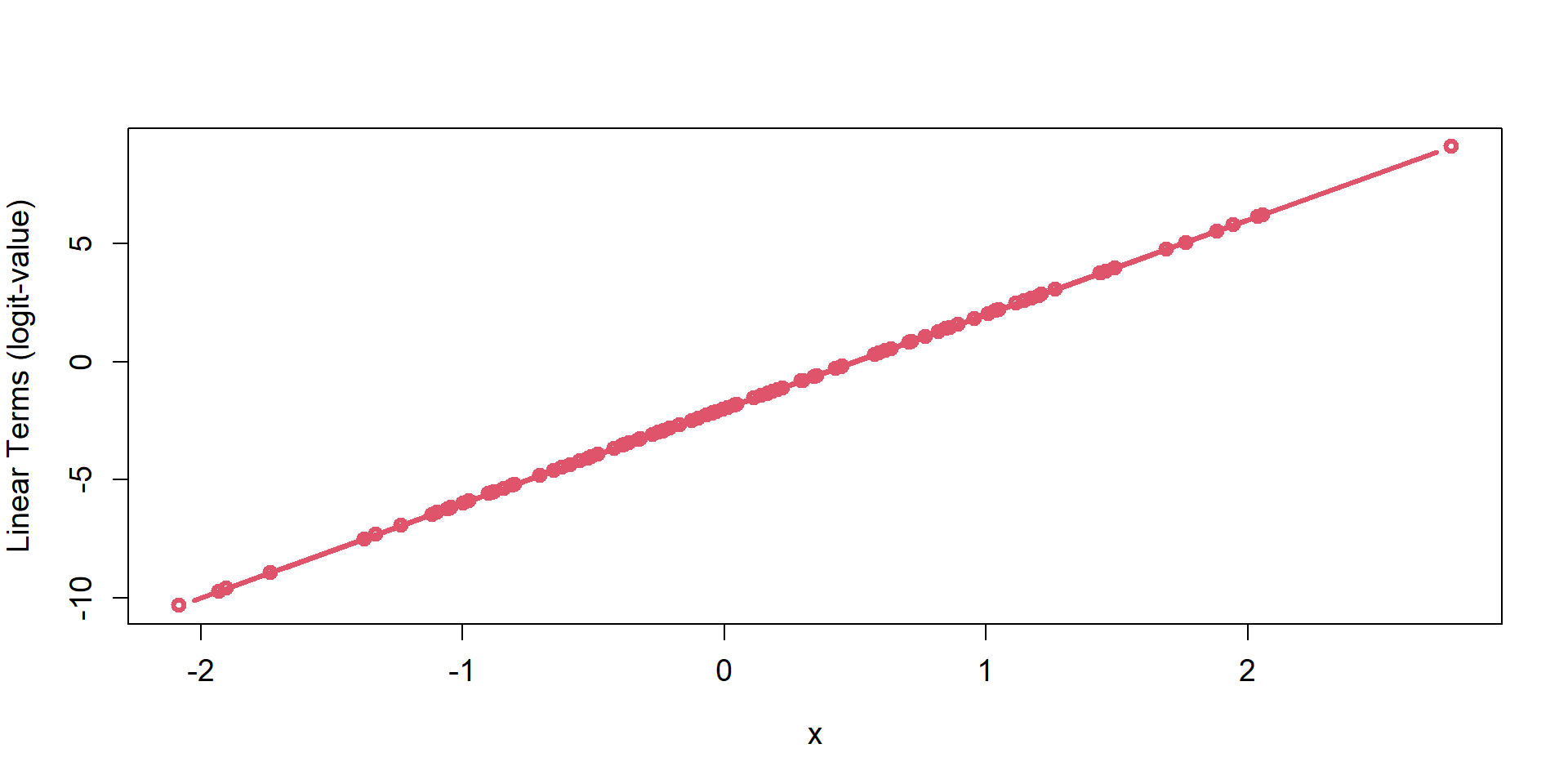[,1]
[1,] 14Generalized Linear Models
Objectives
- GLM framework
- matrix notation
- linear algebra
- design matrix / categorical variable
- glm function
- link functions
- logistic regression
GLM
Generalized linear model framework using matrix notation
\[ \begin{align*} \textbf{y}\sim& [\textbf{y}|\boldsymbol{\mu},\sigma] \\ \text{g}(\boldsymbol{\mu}) =& \textbf{X}\boldsymbol{\beta} \end{align*} \]

Motivation
GLMs:
- t-test
- ANOVA/ANCOVA
- linear regression
- logistic / probit regression
- Poisson regression
- log-linear regression
- survival analysis
- AND MORE!
GLM
Generalized linear model framework
\[ \begin{align*} \textbf{y}\sim& [\textbf{y}|\boldsymbol{\mu},\sigma] \\ \text{g}(\boldsymbol{\mu}) =& \textbf{X}\boldsymbol{\beta} \end{align*} \]
Elements
prob. function to define the RV (\(\textbf{y}\))
parameters or moments of the prob. function (\(\boldsymbol{\mu},\sigma\))
link function (\(\text{g}(\boldsymbol{\mu})\)); deterministic transformation of parameters to new scale
inverse-link function (\(\text{g}^{-1}(\boldsymbol{\textbf{X}\boldsymbol{\beta}})\)); deterministic transformation of linear combination back to parameter scale
design matrix of the explanatory variables (\(\textbf{X}\)); these are known
coefficient parameters (\(\boldsymbol{\beta}\)); needs estimating
Linear Regression
index notation
\[ \begin{align*} y_{i} \sim& \text{Normal}(\mu_{i},\sigma) \\ \mu_{i} =& \beta_{0} + \beta_{1}x_{1i} + \beta_{2}x_{2i} \end{align*} \]
matrix notation
\[ \begin{align*} \textbf{y}\sim& \text{Normal}(\boldsymbol{\mu},\sigma)\\ \boldsymbol{\mu} =& \text{g}^{-1}(\textbf{X}\boldsymbol{\beta})\\ \text{g}(\boldsymbol{\mu}) =& \textbf{X}\boldsymbol{\beta} \end{align*} \]
\[\begin{align*} \textbf{y} = \begin{bmatrix} y_{1} \\ y_{2} \\ y_{3} \\ . \\ . \\ y_{n} \end{bmatrix} \textbf{X} = \begin{bmatrix} 1 & x_{1,2} & x_{1,3} \\ 1 & x_{2,2} & x_{2,3} \\ 1 & x_{3,2} & x_{3,3} \\ . & . .\\ . & . .\\ n & x_{n,2} & x_{n,3} \end{bmatrix} \boldsymbol{\beta} = \begin{bmatrix} \beta_0 \\ \beta_1 \\ \beta_2 \\ \end{bmatrix} \end{align*}\]
n = sample size x\(_{2}\) & x\(_{3}\) are independent variables
Linear Algebra
\(\text{g}(\boldsymbol{\mu}) = \textbf{X}\boldsymbol{\beta}\)
\(\textbf{X}\) is called the Design Matrix.
\(\boldsymbol{\beta}\) is a vector of coefficients.
\[ \textbf{X}= \begin{bmatrix} 1 & x_{1,2} & x_{1,3} \\ 1 & x_{2,2} & x_{2,3} \\ 1 & x_{3,2} & x_{3,3} \end{bmatrix} \boldsymbol{\beta} = \begin{bmatrix} \beta_0 \\ \beta_1 \\ \beta_2 \end{bmatrix} \]
\[ \textbf{X}\boldsymbol{\beta} = \begin{bmatrix} \beta_0\times 1 + \beta_1\times x_{1,2} + \beta_2\times x_{1,3} \\ \beta_0\times 1 + \beta_1\times x_{2,2} + \beta_2\times x_{2,3} \\ \beta_0\times 1 + \beta_1\times x_{3,2} + \beta_2\times x_{3,3} \\ \end{bmatrix}\\ \]
\[ \textbf{X}\boldsymbol{\beta} = \begin{bmatrix} \beta_0 + \beta_1 x_{1,2} + \beta_2 x_{1,3} \\ \beta_0 + \beta_1 x_{2,2} + \beta_2 x_{2,3} \\ \beta_0 + \beta_1 x_{3,2} + \beta_2 x_{3,3} \\ \end{bmatrix}\\ \]
\[ \text{g}(\boldsymbol{\mu}) = \textbf{X}\boldsymbol{\beta} = \begin{bmatrix} \text{lt}_{1} \\ \text{lt}_{2} \\ \text{lt}_{3} \end{bmatrix} \] lt = linear terms
\[ \boldsymbol{\mu} = \text{g}^{-1}(\textbf{X}\boldsymbol{\beta})\ = \textbf{X}\boldsymbol{\beta} / 1= \begin{bmatrix} \text{lt}_{1}/1 \\ \text{lt}_{2}/1 \\ \text{lt}_{3}/1 \end{bmatrix} \]
\[ \boldsymbol{\mu} = \textbf{X}\boldsymbol{\beta} = \begin{bmatrix} \mu_{1} \\ \mu_{2} \\ \mu_{3} \end{bmatrix} \]
Linear Algebra
\(\textbf{y}'\textbf{y}\)
\(\textbf{y}' \cdot \textbf{y}\)
\[ =\begin{bmatrix} 1 & 2 & 3 \end{bmatrix} \begin{bmatrix} 1 \\ 2 \\ 3 \\ \end{bmatrix} \]
\[ =\begin{bmatrix} (1\times1) + (2\times2) + (3\times3)\\ \end{bmatrix} \\= [14] \]
Linear Algebra
When can we do matrix multiplication?
Linear Algebra
Note that
\(\textbf{y}'\textbf{y} \neq \textbf{y}\textbf{y}'\)
Elephant Linear Regression Example
Categorical Variable
weight sex
1 11488.991 Male
2 4105.442 Female
3 4299.308 FemaleElephant Linear Regression Example
Categorical Variable
Call:
glm(formula = weight ~ sex, family = gaussian(link = identity),
data = dat)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4970.9 120.9 41.11 <2e-16 ***
sexMale 6848.1 180.2 37.99 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 1608053)
Null deviance: 2639772182 on 199 degrees of freedom
Residual deviance: 318394574 on 198 degrees of freedom
AIC: 3429.7
Number of Fisher Scoring iterations: 2Elephant Linear Regression Example
Categorical Variable
\(x_{2}\) as an indicator of sex, female (0) or male (1)
\(x_{3}\) elephant age
\[ \textbf{weight} \sim \text{Normal}(\boldsymbol{\mu},\sigma)\\ \\ \boldsymbol{\mu} =\textbf{X}\boldsymbol{\beta} = \begin{bmatrix} \beta_0 + (\beta_1\times 1) + (\beta_2\times 10) \\ \beta_0 + (\beta_1\times 0) + (\beta_2\times 12) \\ \beta_0 + (\beta_1\times 0) + (\beta_2\times 15) \\ \end{bmatrix}\\ \]
\[ \hat{\boldsymbol{\mu}} = \textbf{X}\hat{\boldsymbol{\beta}} = \begin{bmatrix} 2552.82 + (6828.96\times 1) + (145.74\times 10) \\ 2552.82 + (6828.96\times 0) + (145.74\times 12) \\ 2552.82 + (6828.96\times 0) + (145.74\times 15) \\ \end{bmatrix}\\ \]
\[ \hat{\boldsymbol{\mu}} = \textbf{X}\hat{\boldsymbol{\beta}} = \begin{bmatrix} 2552.82 + 6828.96 + 1457.4 \\ 2552.82 + 0 + 1748.88 \\ 2552.82 + 0 + 2186.1 \\ \end{bmatrix}\\ \]
\[ \hat{\boldsymbol{\mu}} = \textbf{X}\hat{\boldsymbol{\beta}} = \begin{bmatrix} 10401.98 \\ 6779.52 \\ 5322.02 \\ \end{bmatrix}\\ \]
So, what does \(\beta_1\) mean?
glm and design matix
(Intercept) sexMale age.years
1 1 1 10
2 1 0 12
3 1 0 15
4 1 0 12
5 1 1 20
6 1 1 4Sex variable is arranged by ‘Dummy Coding’
MLE Estimator with Linear Algebra
\[ \hat{\boldsymbol{\beta}} = (\textbf{X}'\textbf{X})^{-1}\textbf{X}'\textbf{y} \]
X(Intercept) XsexMale Xage.years
2552.8203 6828.9645 145.7483 Link functions
\(\text{g}(\boldsymbol{\mu}) = \textbf{X}\boldsymbol{\beta}\)
\(\boldsymbol{\mu} = \text{g}^{-1}(\textbf{X}\boldsymbol{\beta})\)
Link functions map parameters from one support to another.
Why is that important for us?
To put a linear model on parameters of interest and ensure the parameter support is maintained.
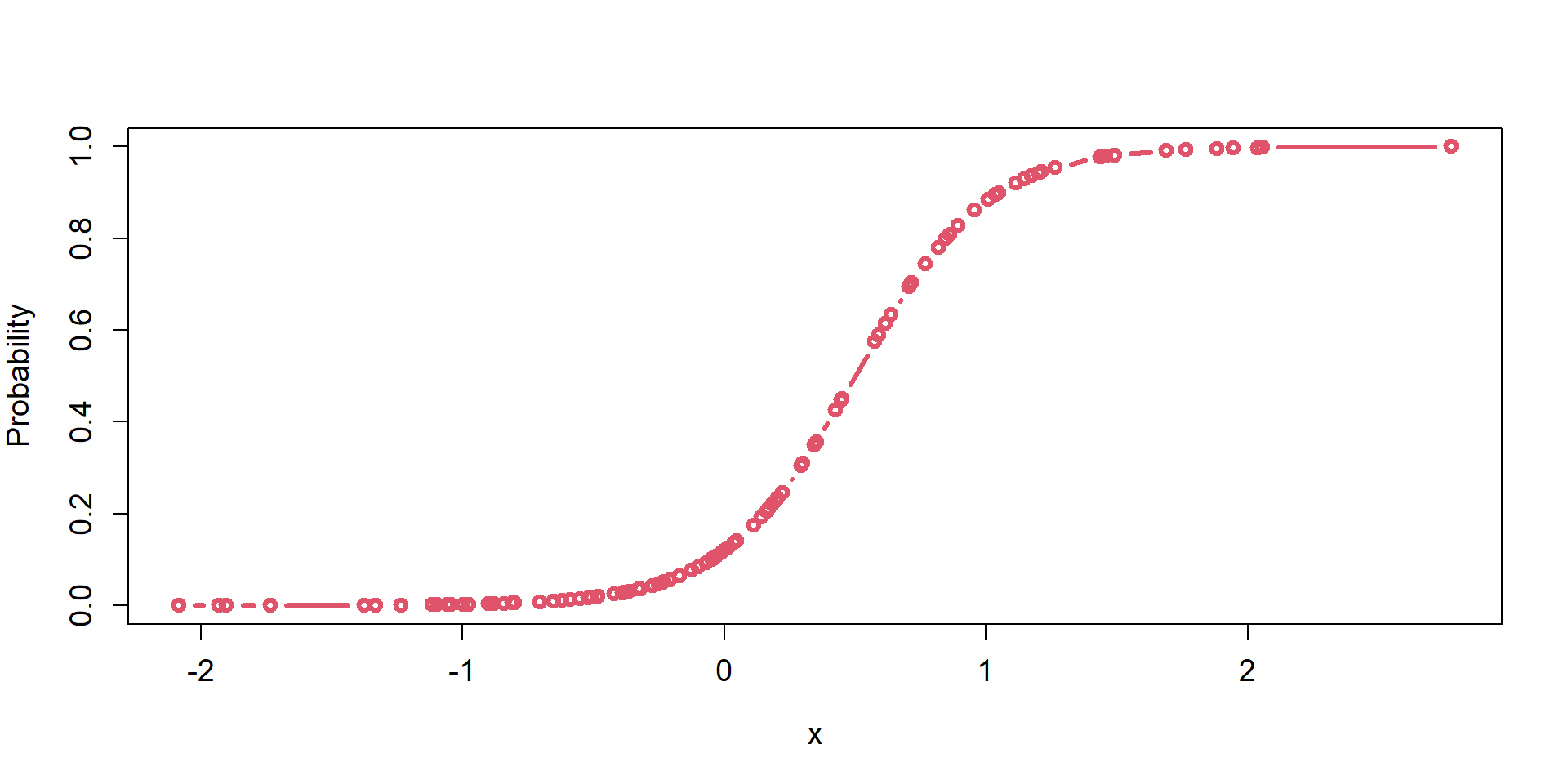
Computers do not like boundaries (e.g., 0 or 1). It’s easier to guess values to evaluate in a maximum liklihood optimization when there are no bounds. (\(-\infty\), \(\infty\))
Link functions
Identity
No transformation is needed because the parameter support is maintained. \[ \begin{align*} \boldsymbol{\mu} \in& (-\infty,\infty)\\ \textbf{X}\boldsymbol{\beta} \in& (-\infty,\infty) \end{align*} \]
A Model by another name
| Model Name | \([y|\boldsymbol{\theta}]\) | Link |
|---|---|---|
| ANOVA (\(x_{1}\) is categorical/multiple levels) | Normal | identity |
| ANCOVA (\(x_{1}\) is categorical, \(x_{2}\) is continuous)) | Normal | identity |
| t-test (\(x_{1}\) is categorical with 2 levels) | Normal | identity |
| Linear Regression \(x_{1}\) is continuous | Normal | identity |
| Multiple Linear Regression \(x_{p}\) is continuous | Normal | identity |
| Logistic Regression | Binomial | logit |
| Probit Regression | Binomial | probit |
| Log-linear Regression | Poisson | log |
| Poisson Regression | Poisson | log |
| Survival Analysis | Exponential | log |
| Inverse Polynomial | Gamma | Reciprocal |
Logistic Regression (logit link)
\[ \begin{align*} \textbf{y} \sim& \text{Binomial}(N,\boldsymbol{p}) \end{align*} \]
\[ \begin{align*} \text{g}(\boldsymbol{p}) =& \text{logit}(\boldsymbol{p}) = \text{log}(\frac{\boldsymbol{p}}{1-\boldsymbol{p}}) \end{align*} \]
inverse-logit (expit)
\[ \boldsymbol{p} = g^{-1}(\boldsymbol{\textbf{X}\boldsymbol{\beta}}) = \text{logit}^{-1}(\textbf{X}\boldsymbol{\beta}) = \frac{e^{\textbf{X}\boldsymbol{\beta}}}{e^{\textbf{X}\boldsymbol{\beta}}+1} \]
Logistic Regression
Full model
\[ \begin{align*} \textbf{y} \sim& \text{Binomial}(N,\boldsymbol{p})\\ \text{logit}(\boldsymbol{p}) =& \textbf{X}\boldsymbol{\beta} \end{align*} \]
Remember,
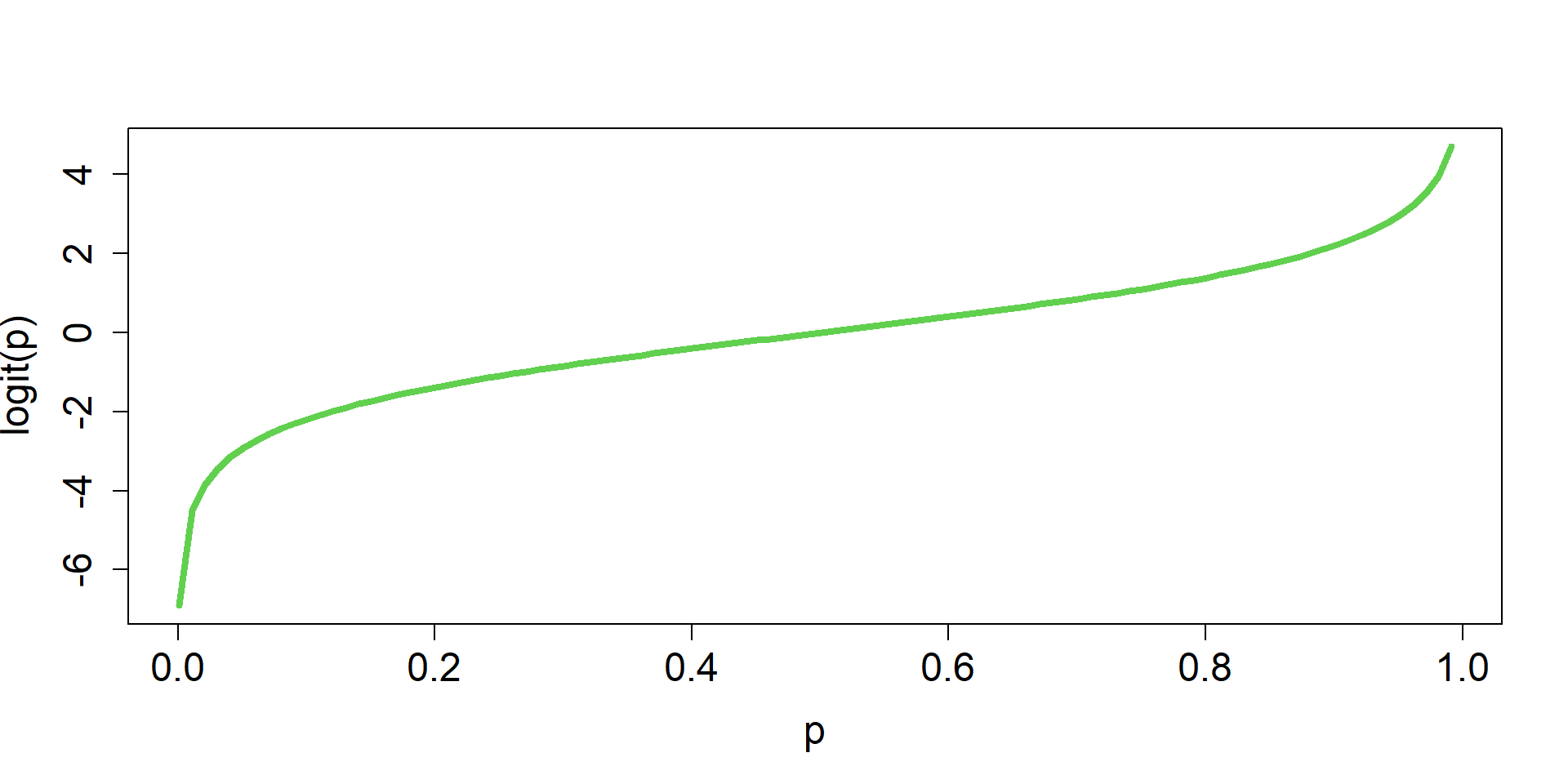
\(\boldsymbol{p} \in [0,1]\)
\(\textbf{X}\boldsymbol{\beta} \in (-\infty,\infty)\)
logit/p mapping
Logistic Regression Simulation
# marginal coefficients (on logit-scale)
beta=c(-2,4)
#linear terms
lt = X%*%beta
#transformation via link function to probability scale
p=plogis(lt)
head(round(p,digits=2)) [,1]
1 0.98
2 0.02
3 0.90
4 0.01
5 0.95
6 0.00 [1] 1 0 1 0 1 0 0 1 0 1 0 0 0 1 1 0 0 1 1 0 0 1 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0
[38] 0 1 1 0 1 0 1 0 1 1 0 1 1 1 1 0 0 1 1 0 1 0 1 0 1 1 0 0 0 0 0 0 0 0 0 0 0
[75] 0 0 0 0 0 1 1 0 0 1 0 0 0 0 1 1 0 0 1 0 0 0 0 0 1 0Logistic Regression Estimation
Call:
glm(formula = y ~ 0 + X, family = binomial(link = "logit"))
Coefficients:
Estimate Std. Error z value Pr(>|z|)
X(Intercept) -2.5304 0.6473 -3.909 9.25e-05 ***
XVar1 5.4198 1.2232 4.431 9.38e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 138.63 on 100 degrees of freedom
Residual deviance: 39.14 on 98 degrees of freedom
AIC: 43.14
Number of Fisher Scoring iterations: 7Logistic Regression Evaluation
[1] 100 1000 #Estimate coefs for all 100 samples
coef.est=apply(y.many,2,FUN=function(y){
model1=glm(y~0+X, family = binomial(link = "logit"))
model1$coefficients
})
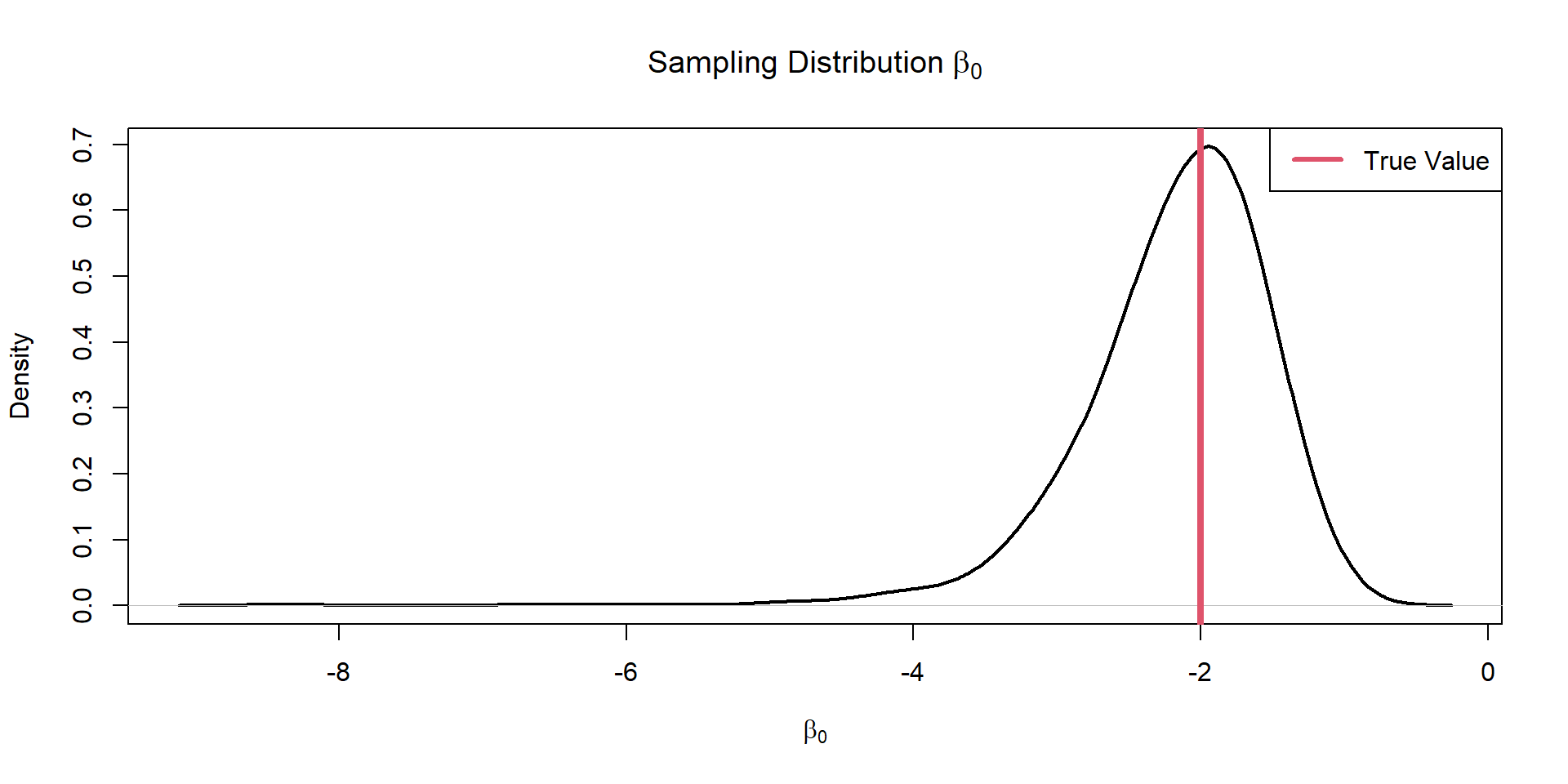
dim(coef.est)[1] 2 1000Plot Intercept
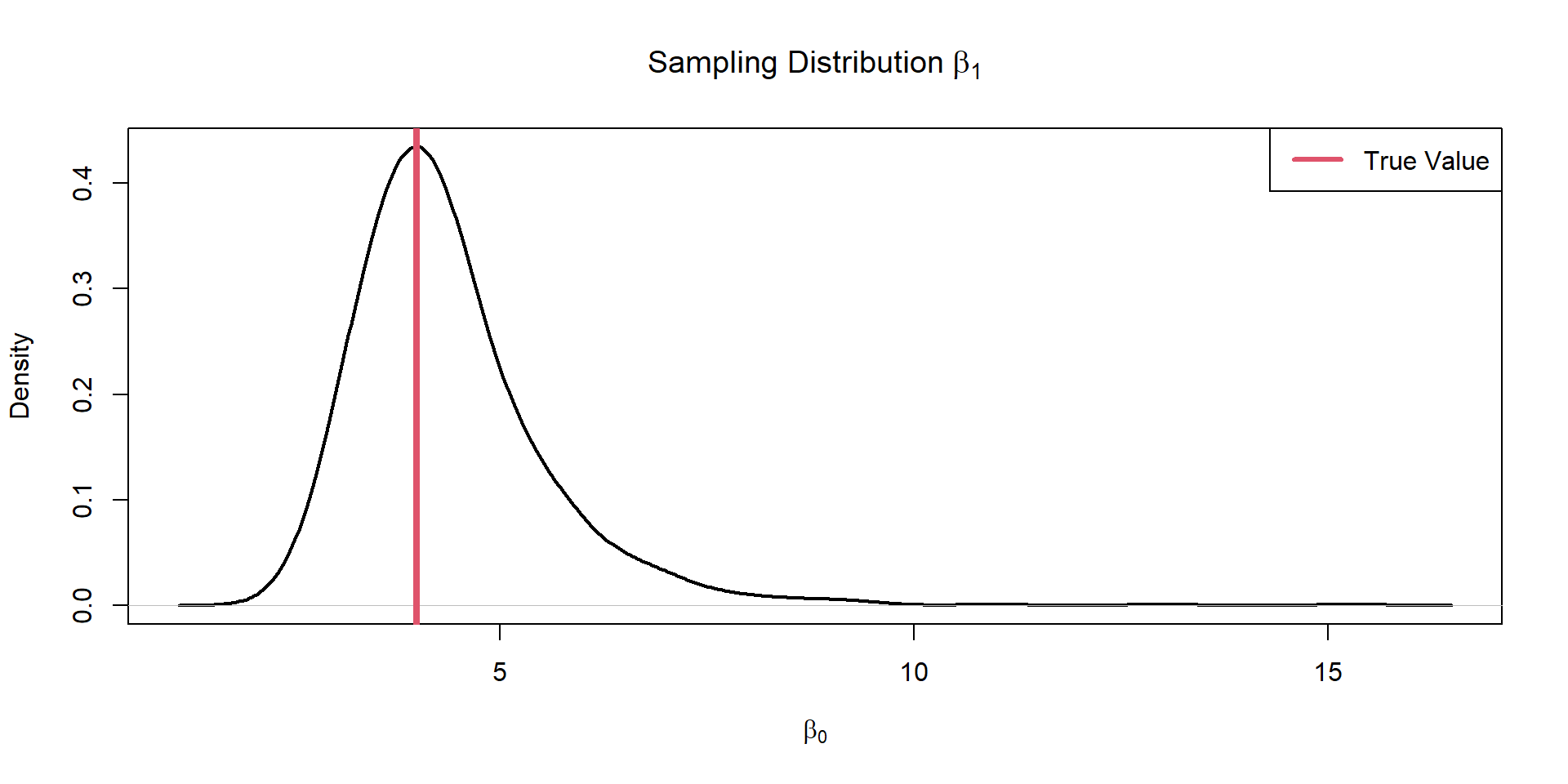
Plot Slope
Evaluate Sampling Distributions
Is our model biased?
Evaluate Sampling Distributions
Are we going to estimate the sign of the coef correctly?
Logistic Regression Biased Correction
brglm: Bias Reduction in Binomial-Response Generalized Linear Models
Recap
- GLM framework: What are key components?
- matrix notation - why learn this?
- link functions - what are they?