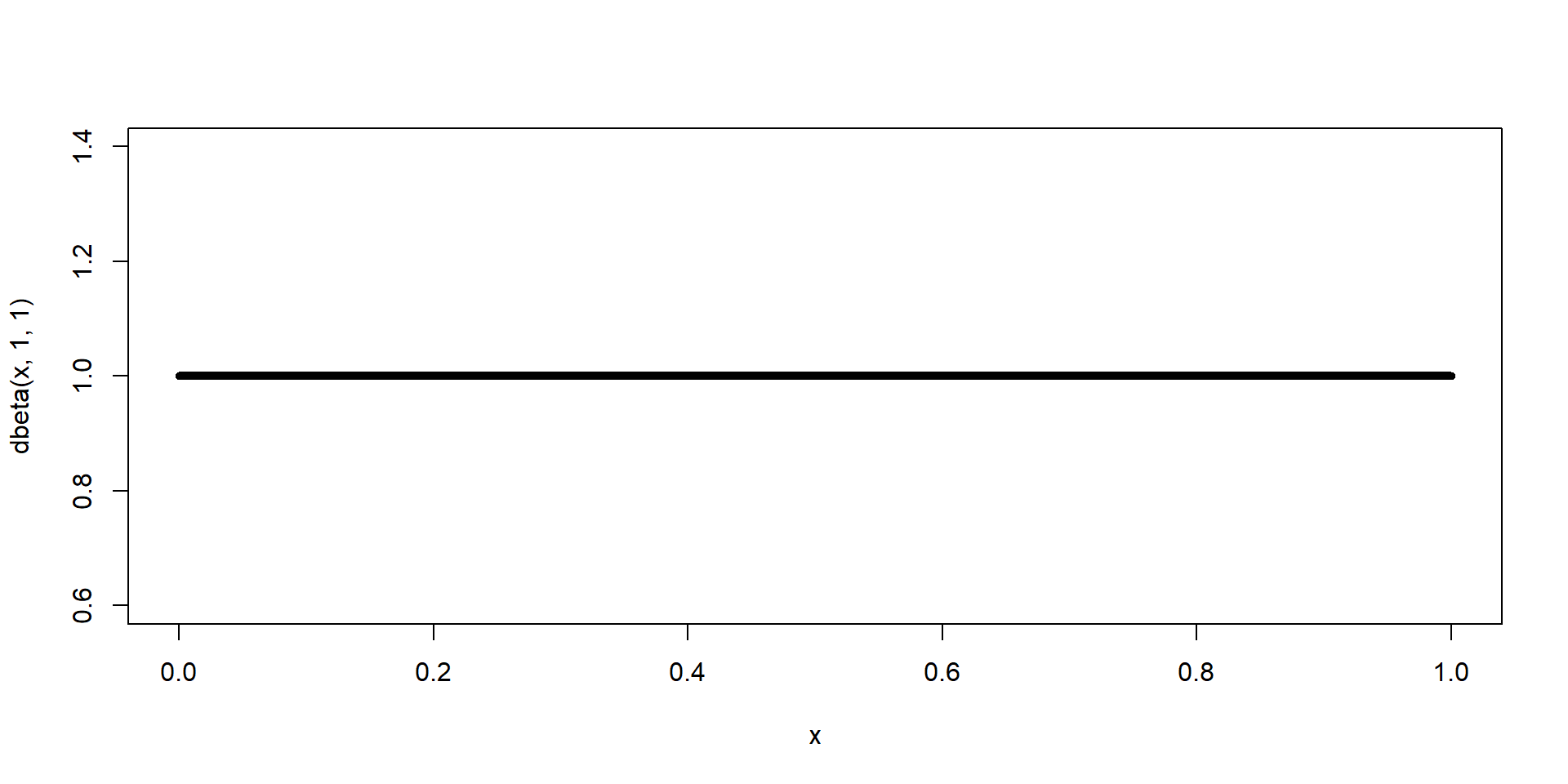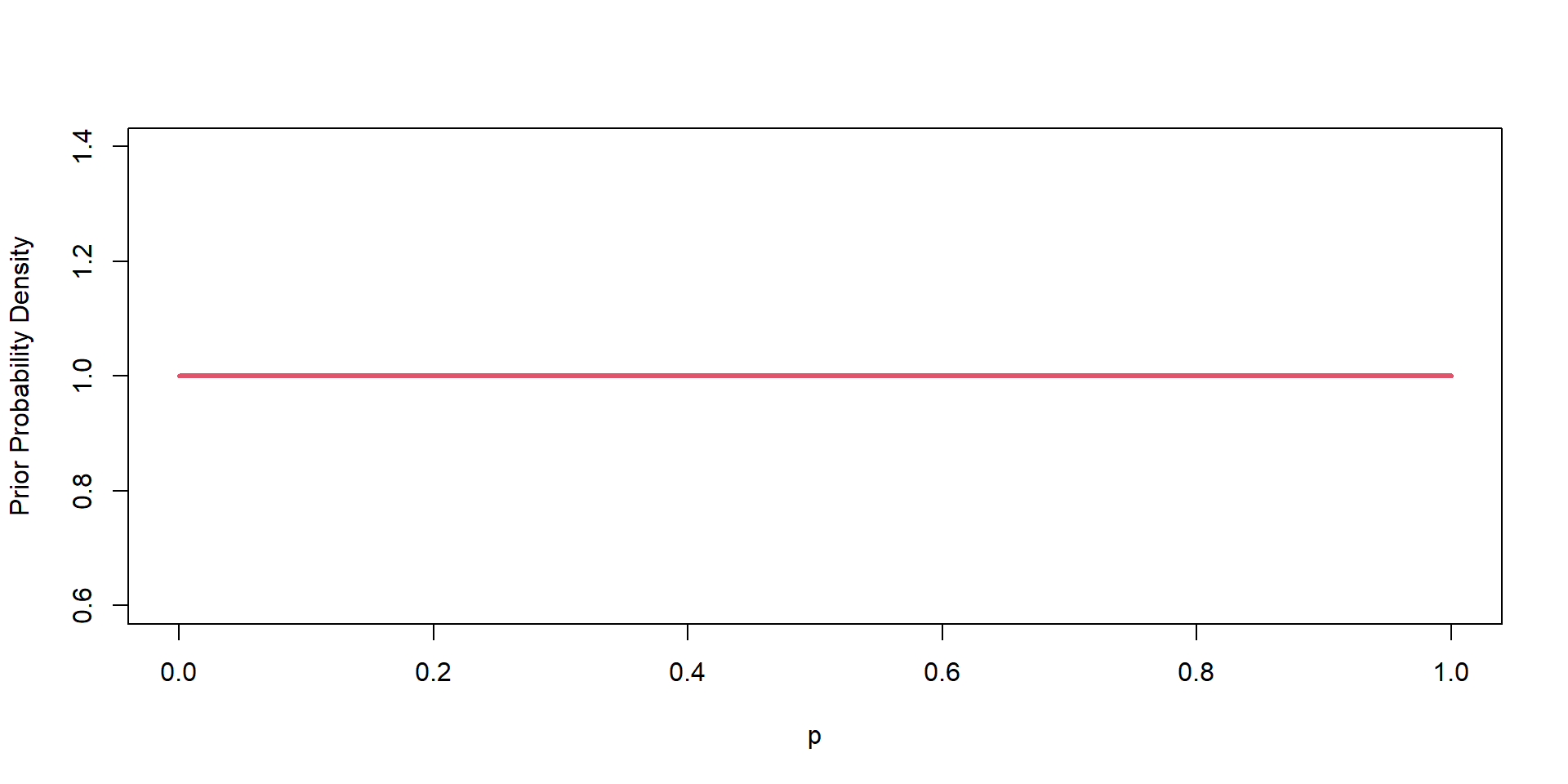[1] 102.67671 81.11546 81.75260 87.77246 73.99043 80.70631 76.26219
[8] 83.99927 64.74208 26.93133
Bayes the way
All things Bayesian
Bayesian Inference
Bayes Thereom
Bayesian Components
- likelihood, prior, evidence, posterior
Conjugacy
Hippo Case Study
Bayesian Computation
Probability, Data, and Parameters
What do we want our model to tell us?
Do we want to make probability statements about our data?
Likelihood = P(data|parameters)
90% CI: the long-run proportion of corresponding CIs that will contain the true value 90% of the time.
Probability, Data, and Parameters
What do we want our model to tell us?
Do we want to make probability statements about our parameters?
Posterior = P(parameters|data)
Alternative Interval: 90% probability that the true value lies within the interval, given the evidence from the observed data.
Likelihood Inference
Estimate of the population size of hedgehogs at two sites.
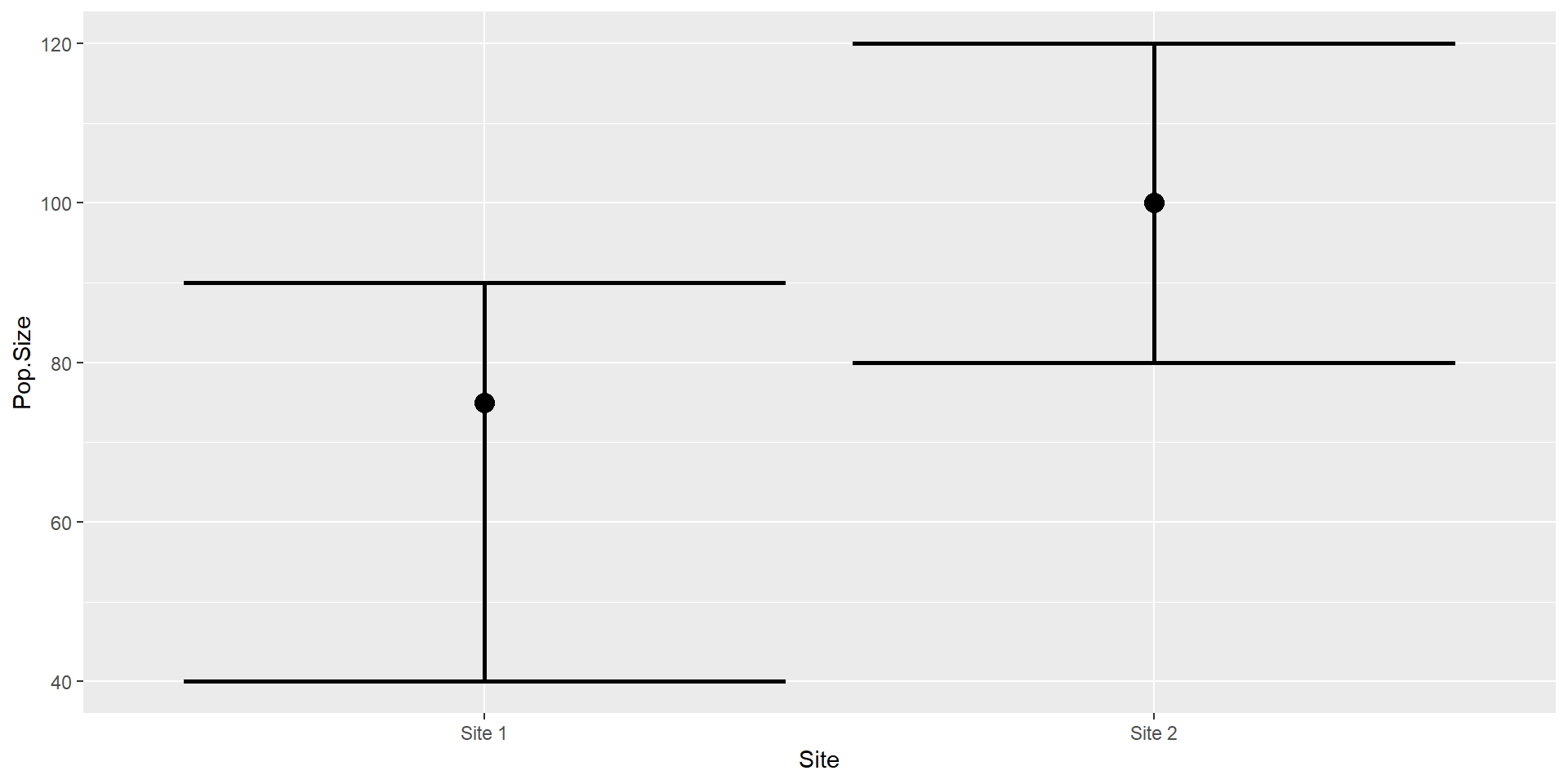
Bayesian Inference
Posterior Samples
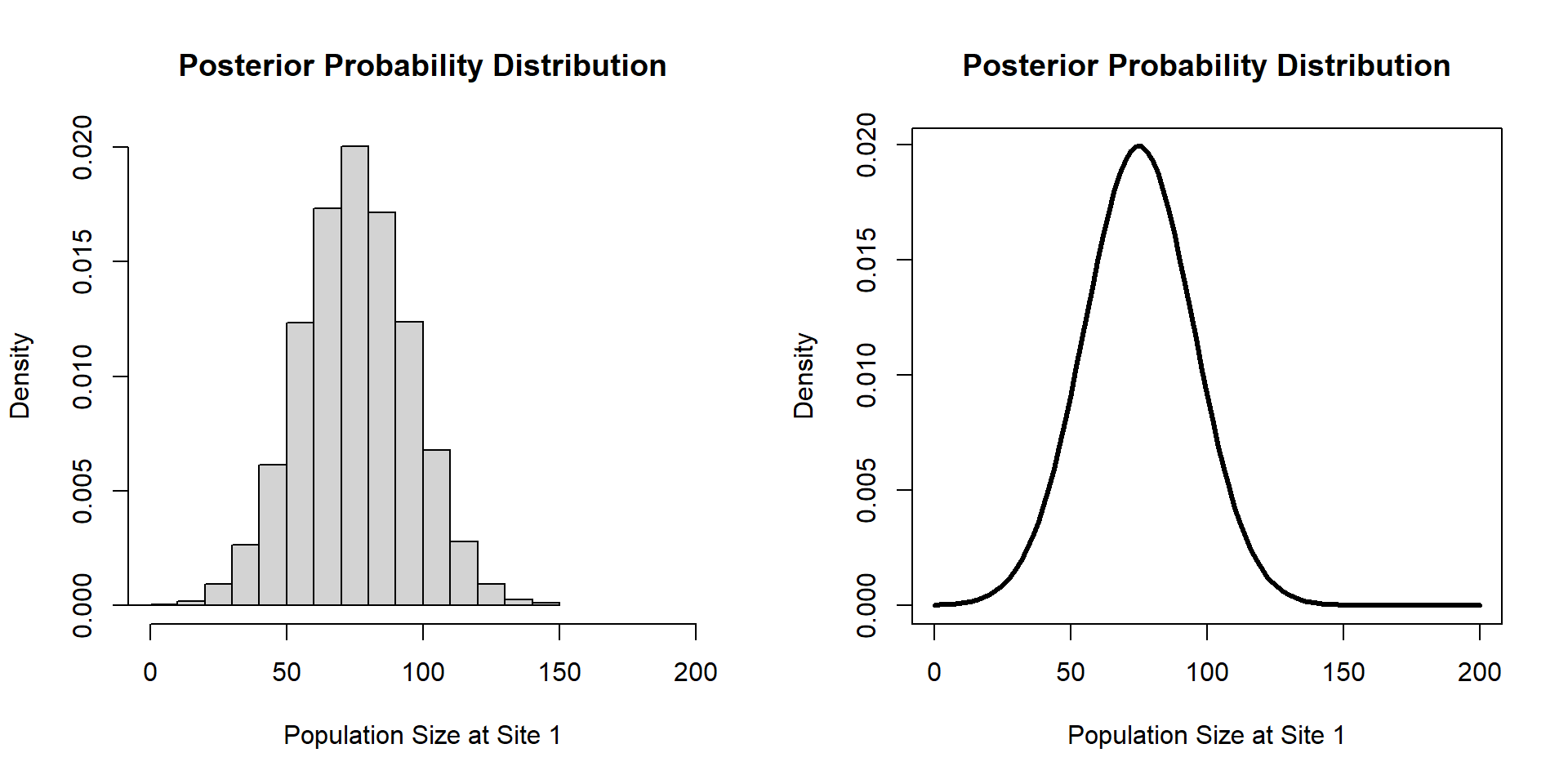
Bayesian Inference
Posterior Samples
[1] 102.67671 81.11546 81.75260 87.77246 73.99043 80.70631 76.26219
[8] 83.99927 64.74208 26.93133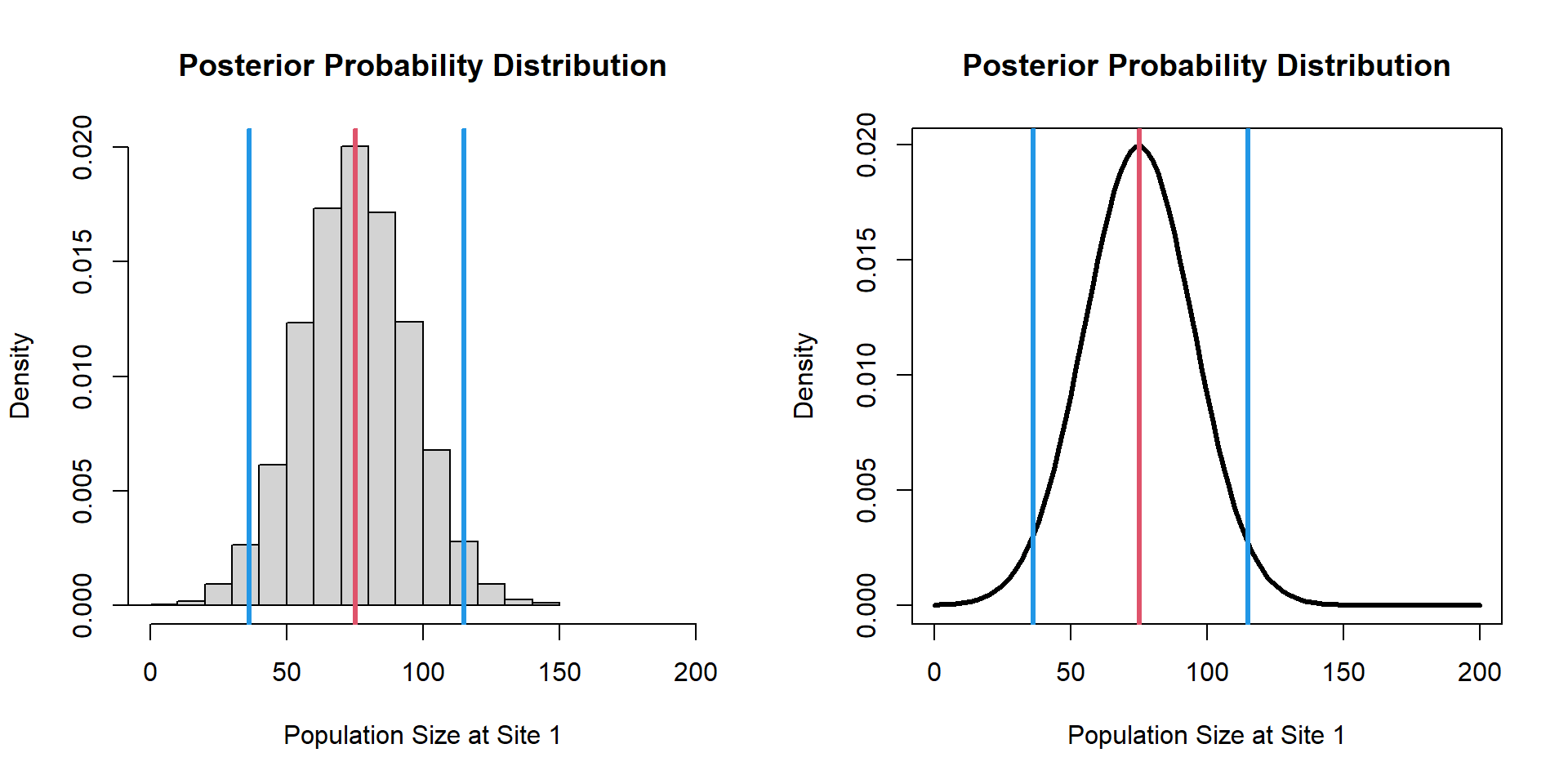
Bayesian Inference
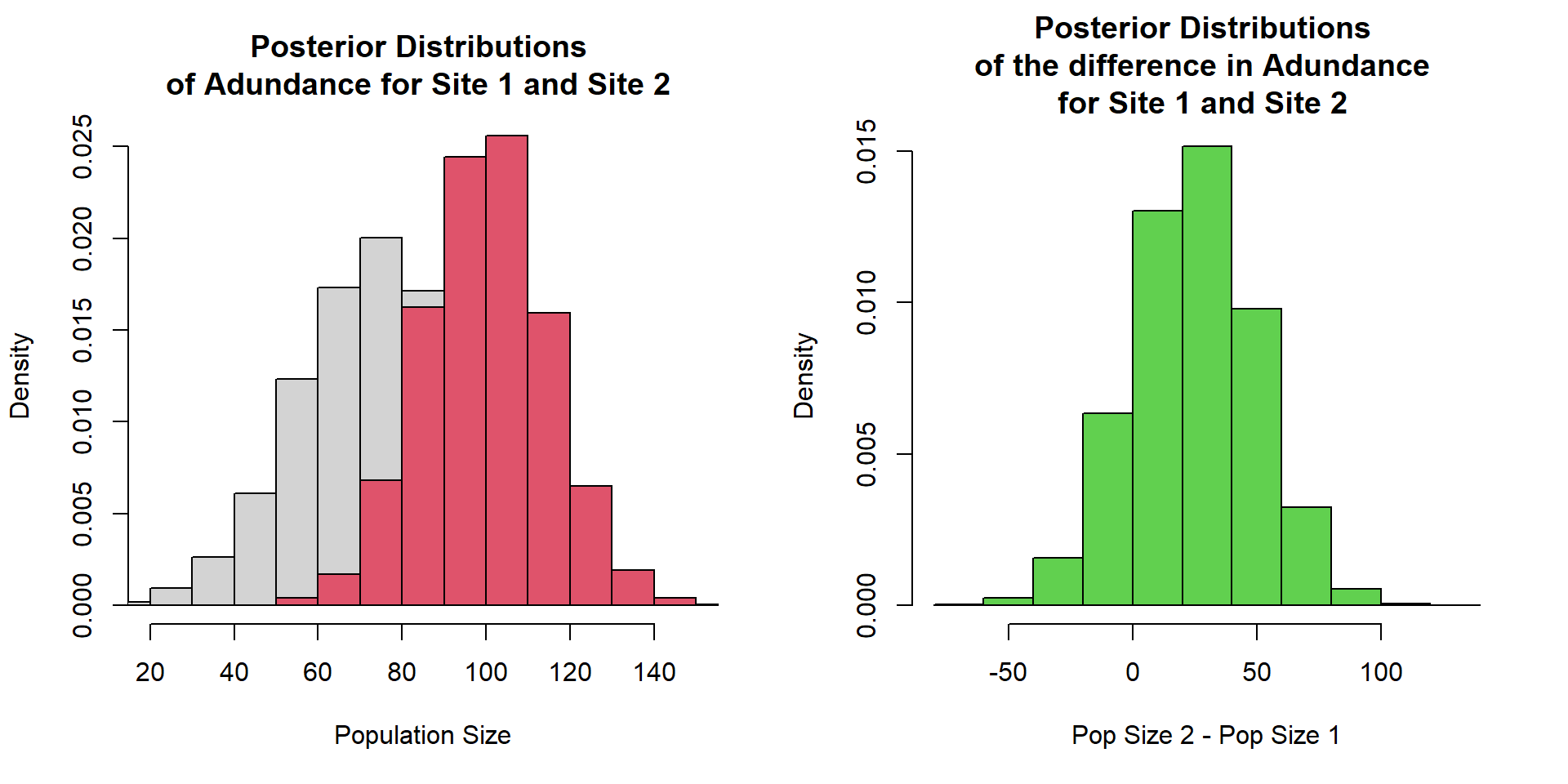
Likelihood Inference (coeficient)
y is Body size of a beetle species
x is elevation
Call:
glm(formula = y ~ x)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.9862 0.2435 4.049 0.000684 ***
x 0.5089 0.4022 1.265 0.221093
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 1.245548)
Null deviance: 25.659 on 20 degrees of freedom
Residual deviance: 23.665 on 19 degrees of freedom
AIC: 68.105
Number of Fisher Scoring iterations: 2Bayesian Inference (coeficient)
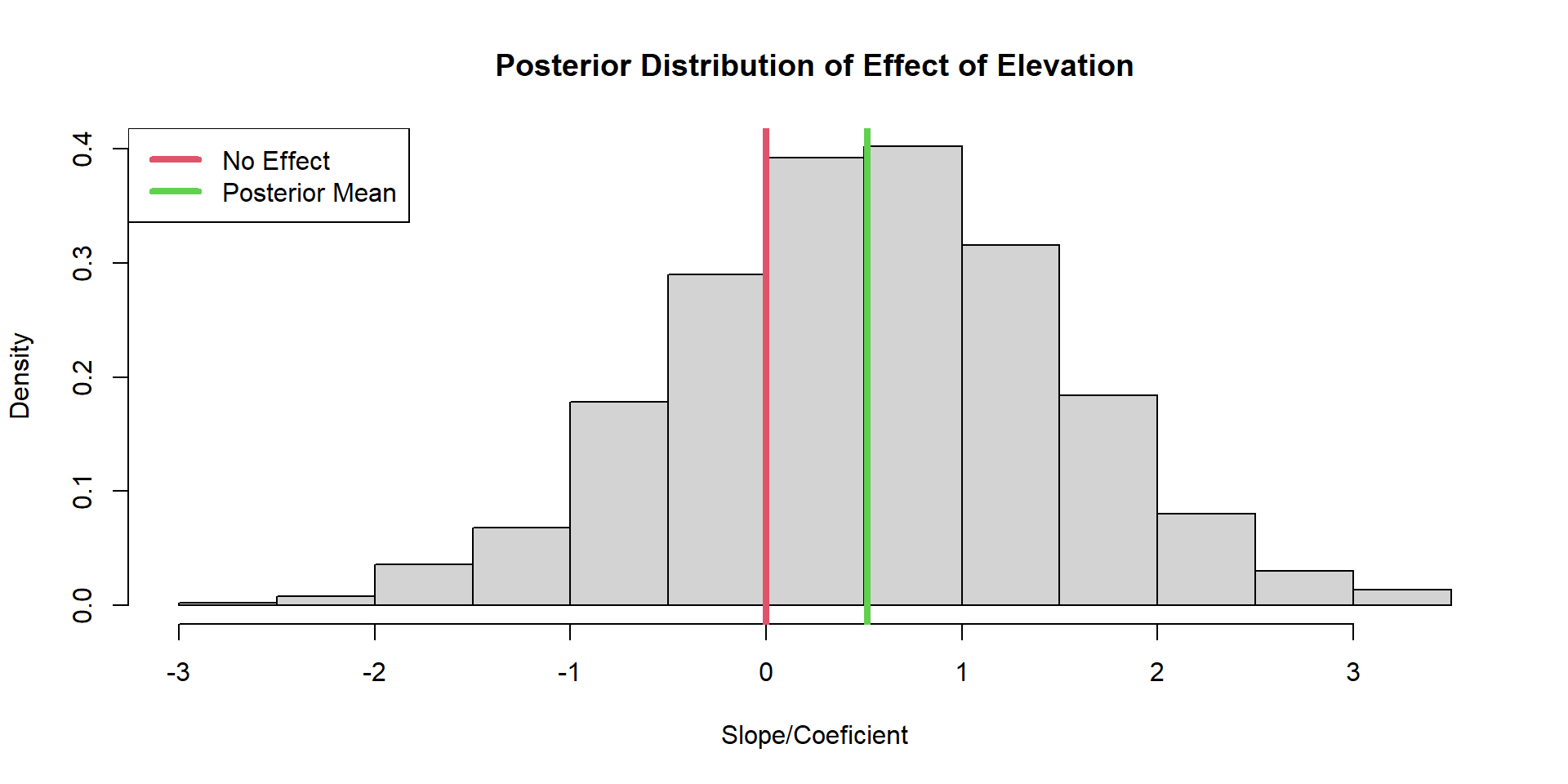
Bayesian Inference (coeficient)
Bayes Theorem
Bayes Theorem
Bayes Theorem
Bayes Theorem
Notice that…
\[ \begin{equation} P(A \cap B) = 0.2 \\ P(A|B)P(B) = 0.4 \times 0.5 = 0.2 \\ P(B|A)P(A) = 0.6666 \times 0.3 = 0.2 \\ \end{equation} \]
\[ \begin{equation} P(A|B)P(B) = P(A \cap B) \\ P(B|A)P(A) = P(A \cap B) \\ \end{equation} \]
\[ \begin{equation} P(B|A)P(A) = P(A|B)P(B) \end{equation} \]
Bayes Theoreom
\[ \begin{equation} P(B|A) = \frac{P(A|B)P(B)}{P(A)} \\ \end{equation} \]
\[ \begin{equation} P(B|A) = \frac{P(A \cap B)}{P(A)} \\ \end{equation} \]
Bayes Components
param = parameters \[ \begin{equation} P(\text{param}|\text{data}) = \frac{P(\text{data}|\text{param})P(\text{param})}{P(\text{data})} \\ \end{equation} \]
Posterior Probability/Belief
Likelihood
Prior Probability
Evidence or Marginal Likelihood
Bayes Components
param = parameters
\[ \begin{equation} P(\text{param}|\text{data}) = \frac{P(\text{data}|\text{param})P(\text{param})}{\int_{\forall \text{ Param}} P(\text{data}|\text{param})P(\text{param})} \end{equation} \]
Posterior Probability/Belief
Likelihood
Prior Probability
Evidence or Marginal Likelihood
Bayes Components
\[ \begin{equation} \text{Posterior} = \frac{\text{Likelihood} \times \text{Prior}}{\text{Evidence}} \\ \end{equation} \]
\[ \begin{equation} \text{Posterior} \propto \text{Likelihood} \times \text{Prior} \end{equation} \]
\[ \begin{equation} \text{Posterior} \propto \text{Likelihood} \end{equation} \]
The Prior
All parameters in a Bayesian model require a prior specified; parameters are random variables.
\(y_{i} \sim \text{Binom}(N, \theta)\)
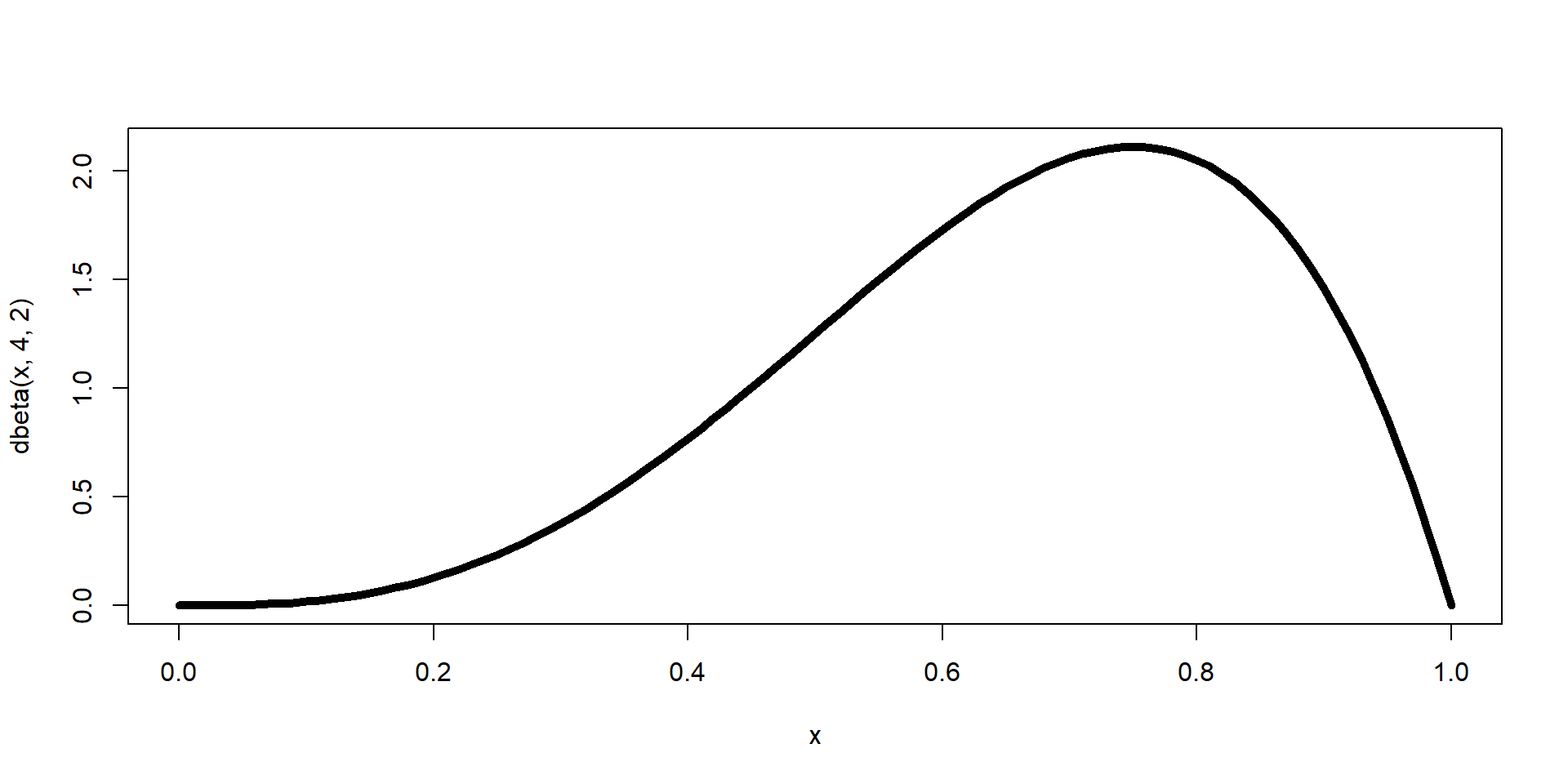
\(\theta \sim \text{Beta}(\alpha = 4, \beta=2)\)
The Prior
\(\theta \sim \text{Beta}(\alpha = 1, \beta=1)\)
The Prior
The prior describes what we know about the parameter before we collect any data
Priors can contain a lot of information (informative priors ) or very little (diffuse priors )
Well-constructed priors can also improve the behavior of our models (computational advantage)
No such thing as a ‘non-informative’ prior
The Prior
Use diffuse priors as a starting point
It’s fine to use diffuse priors as you develop your model but you should always prefer to use “appropriate, well-contructed informative priors” (Hobbs & Hooten, 2015)
The Prior
Use your “domain knowledge”
We can often come up with weakly informative priors just by knowing something about the range of plausible values of our parameters.
The Prior
Dive into the literature
Find published estimates and use moment matching and other methods to convert published estimates into prior distributions
The Prior
Visualize your prior distribution
Be sure to look at the prior in terms of the parameters you want to make inferences about (use simulation!)
The Prior
Do a sensitivity analysis
Does changing the prior change your posterior inference? If not, don’t sweat it. If it does, you’ll need to return to point 2 and justify your prior choice
Bayesian Model
Model
\[ \textbf{y} \sim \text{Bernoulli}(p) \]
Conjugate Distribution
Likelihood (Joint Probability of y)
\[ \mathscr{L}(p|y) = \prod_{i=1}^{n} P(y_{i}|p) = \prod_{i=1}^{N}(p^{y}(1-p)^{1-y_{i}}) \]
Prior Distribution
\[ P(p) = \frac{p^{\alpha-1}(1-p)^{\beta-1}}{B(\alpha,\beta)} \]
Posterior Distribution of p
\[ P(p|y) = \frac{\prod_{i=1}^{N}(p^{y}(1-p)^{1-y_{i}}) \times \frac{p^{\alpha-1}(1-p)^{\beta-1}}{B(\alpha,\beta)} }{\int_{p}(\text{numerator})} \]
\[ P(p|y) \sim \text{Beta}(\alpha^*,\beta^*) \]
\(\alpha^*\) and \(\beta^*\) are called Posterior hyperparameters
\[ \alpha^* = \alpha + \sum_{i=1}^{N}y_i \\ \beta^* = \beta + N - \sum_{i=1}^{N}y_i \\ \]
Hippos
We do a small study on hippo survival and get these data…

Hippos: Likelihood Model
\[ \begin{align*} \textbf{y} \sim& \text{Binomial}(N,p)\\ \end{align*} \]
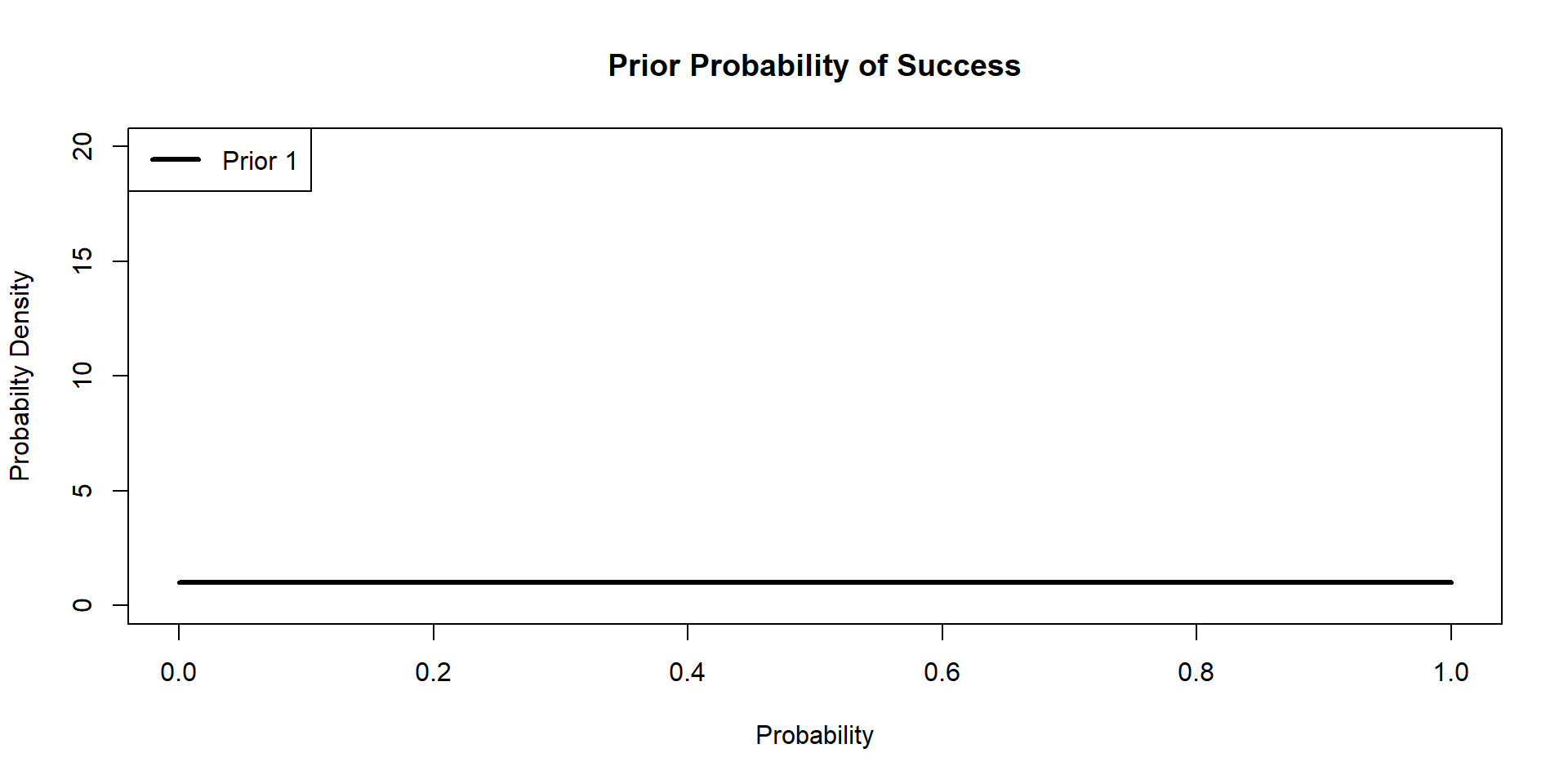
Hippos: Bayesian Model (Prior 1)
\[ \begin{align*} \textbf{y} \sim& \text{Binomial}(N,p)\\ p \sim& \text{Beta}(\alpha,\beta) \end{align*} \]
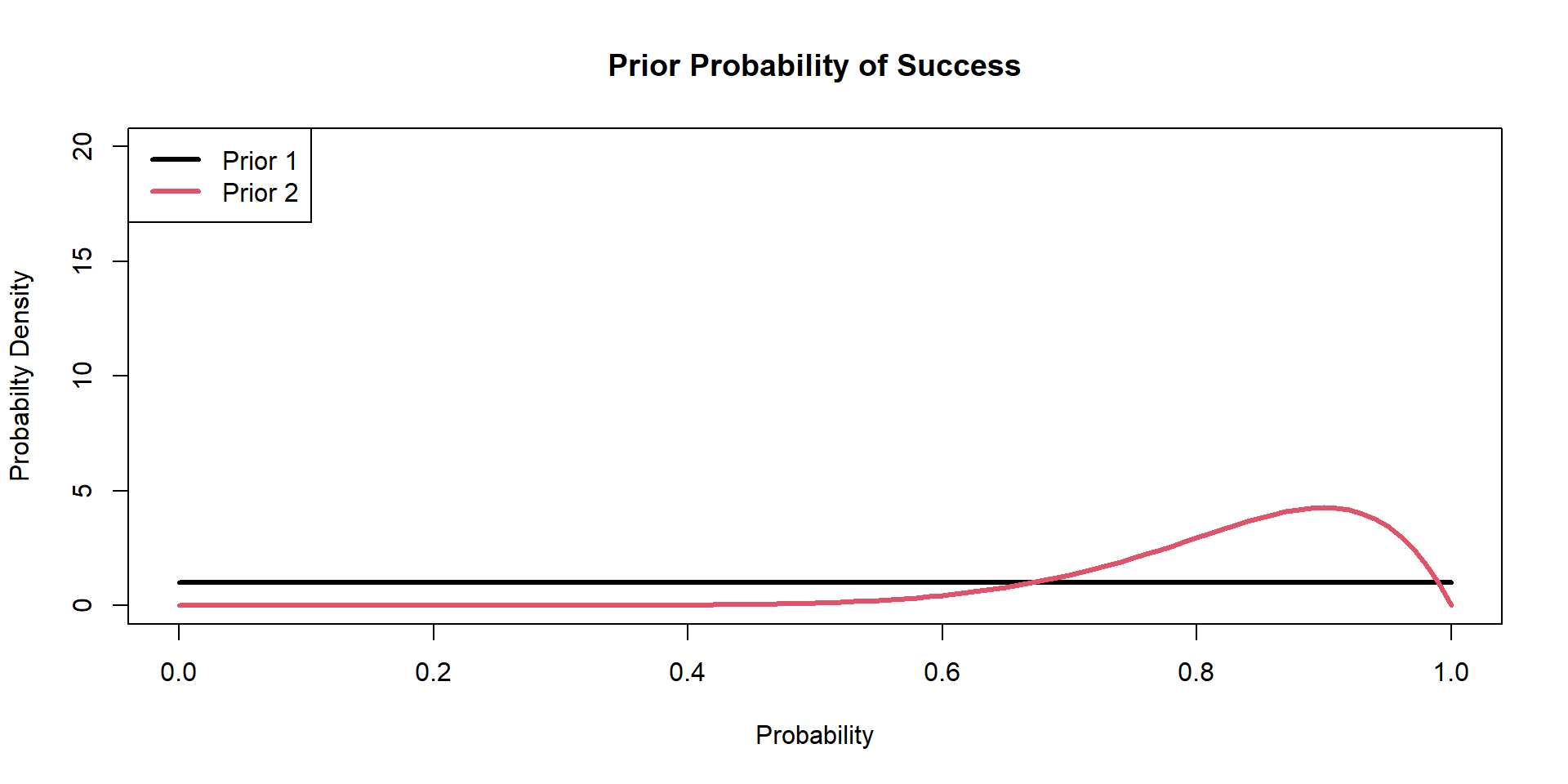
Hippos: Bayesian Model (Prior 2)
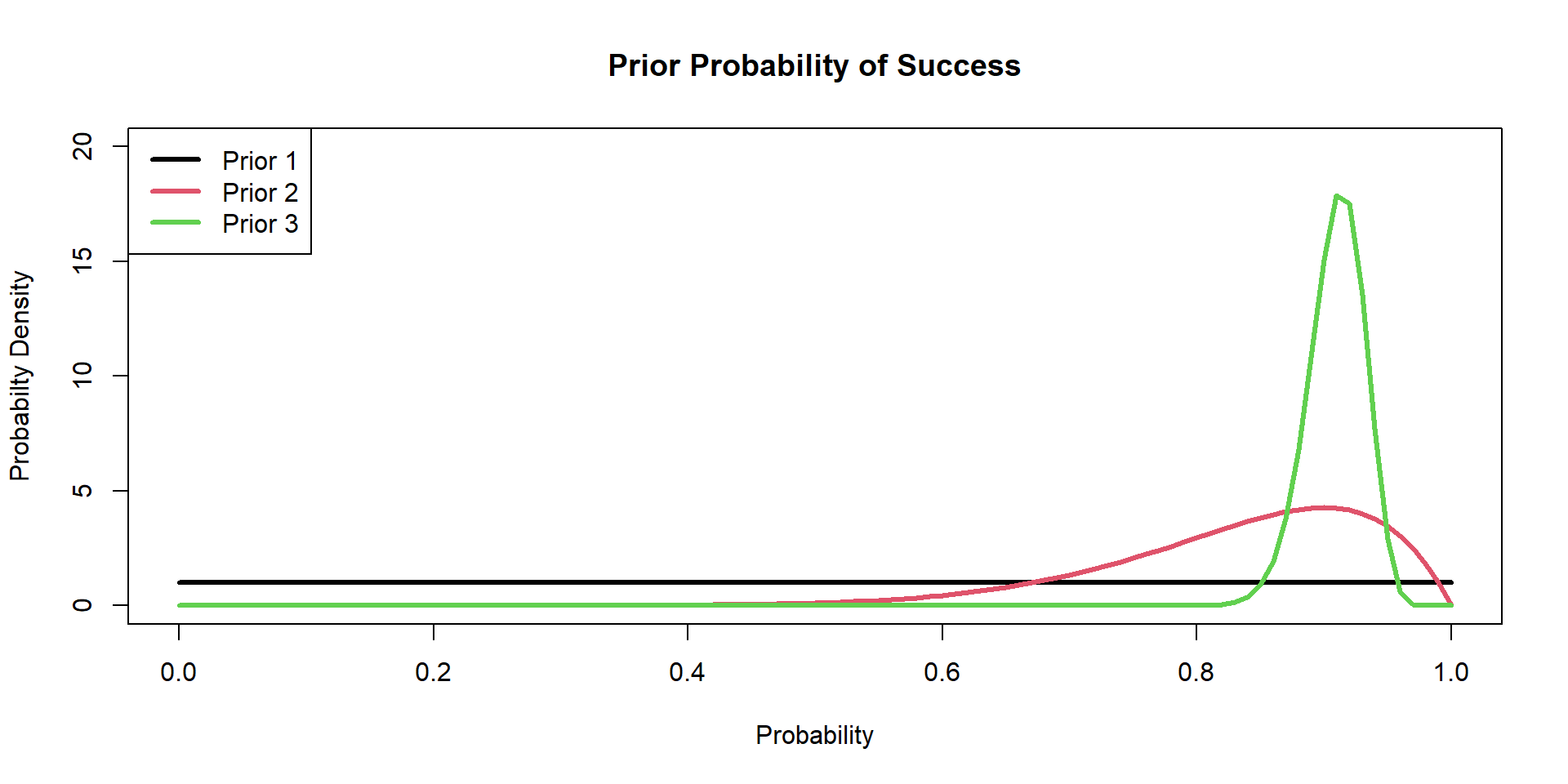
Hippos: Bayesian Model (Prior 3)
Hippos: Bayesian Model (Posteriors)
\[ P(p|y) \sim \text{Beta}(\alpha^*,\beta^*)\\ \alpha^* = \alpha + \sum_{i=1}^{N}y_i \\ \beta^* = \beta + N - \sum_{i=1}^{N}y_i \\ \]
Hippos: Bayesian Model (Posteriors)
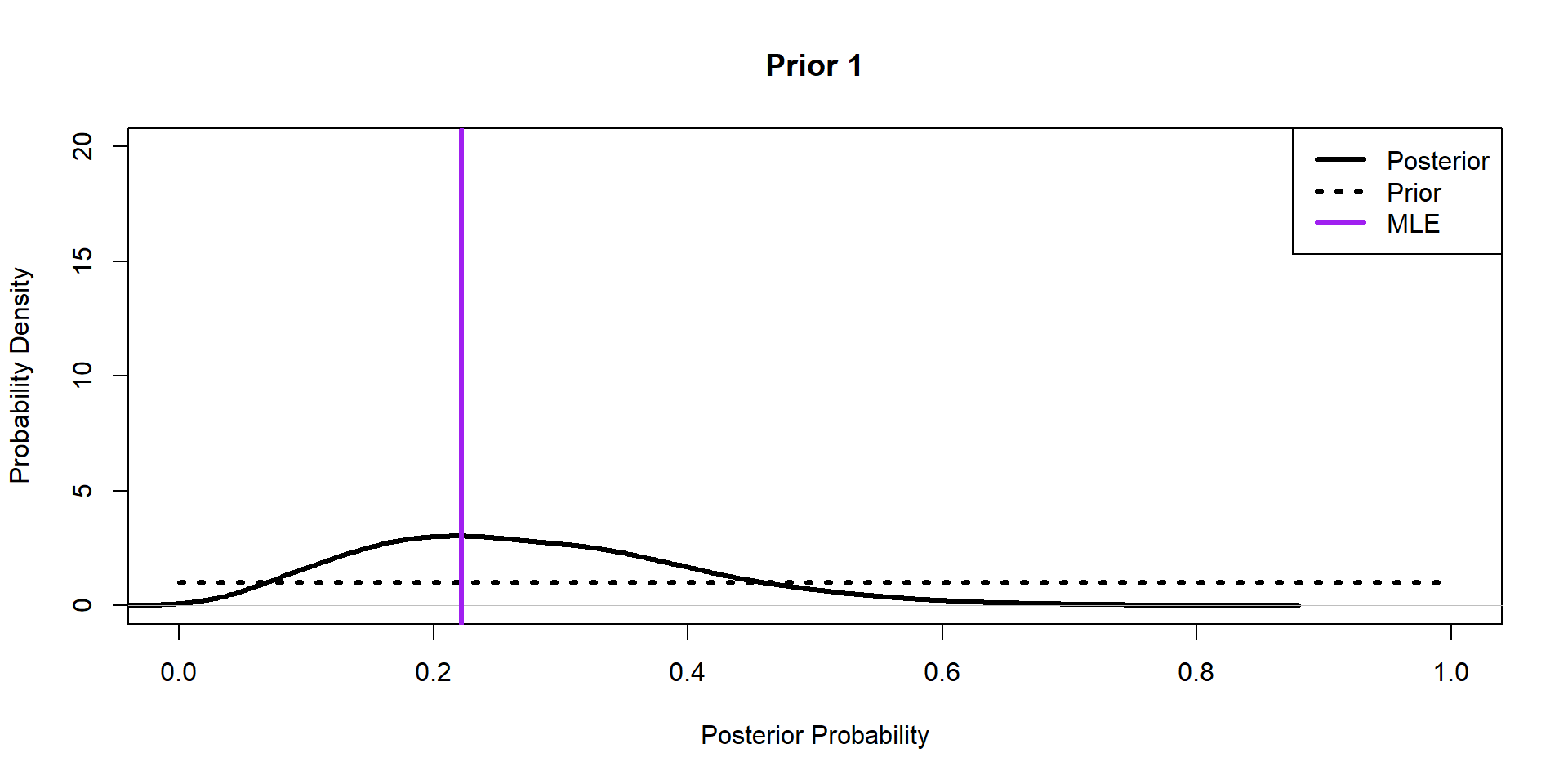
Hippos: Bayesian Model (Posteriors)
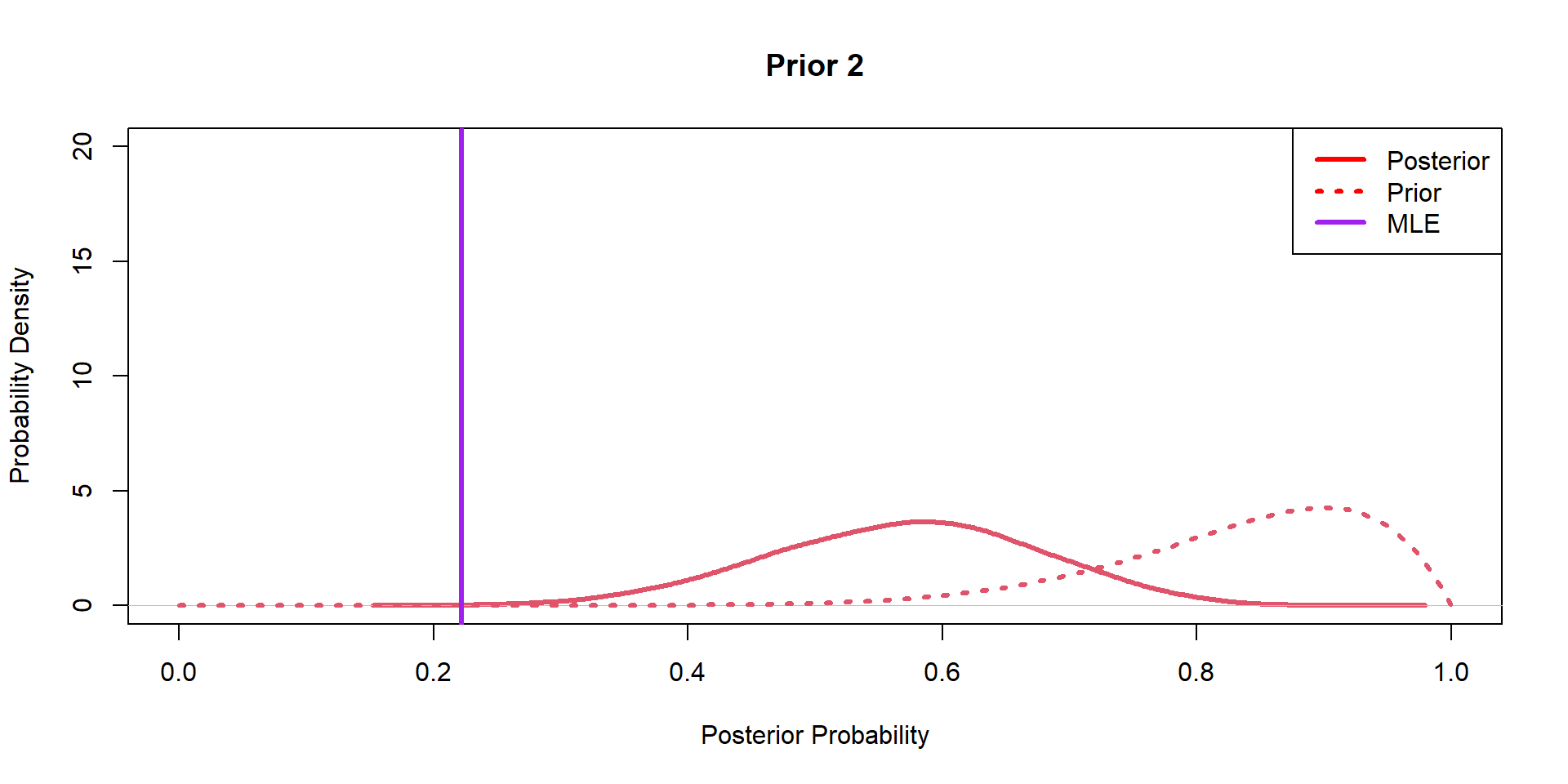
Hippos: Bayesian Model (Posteriors)
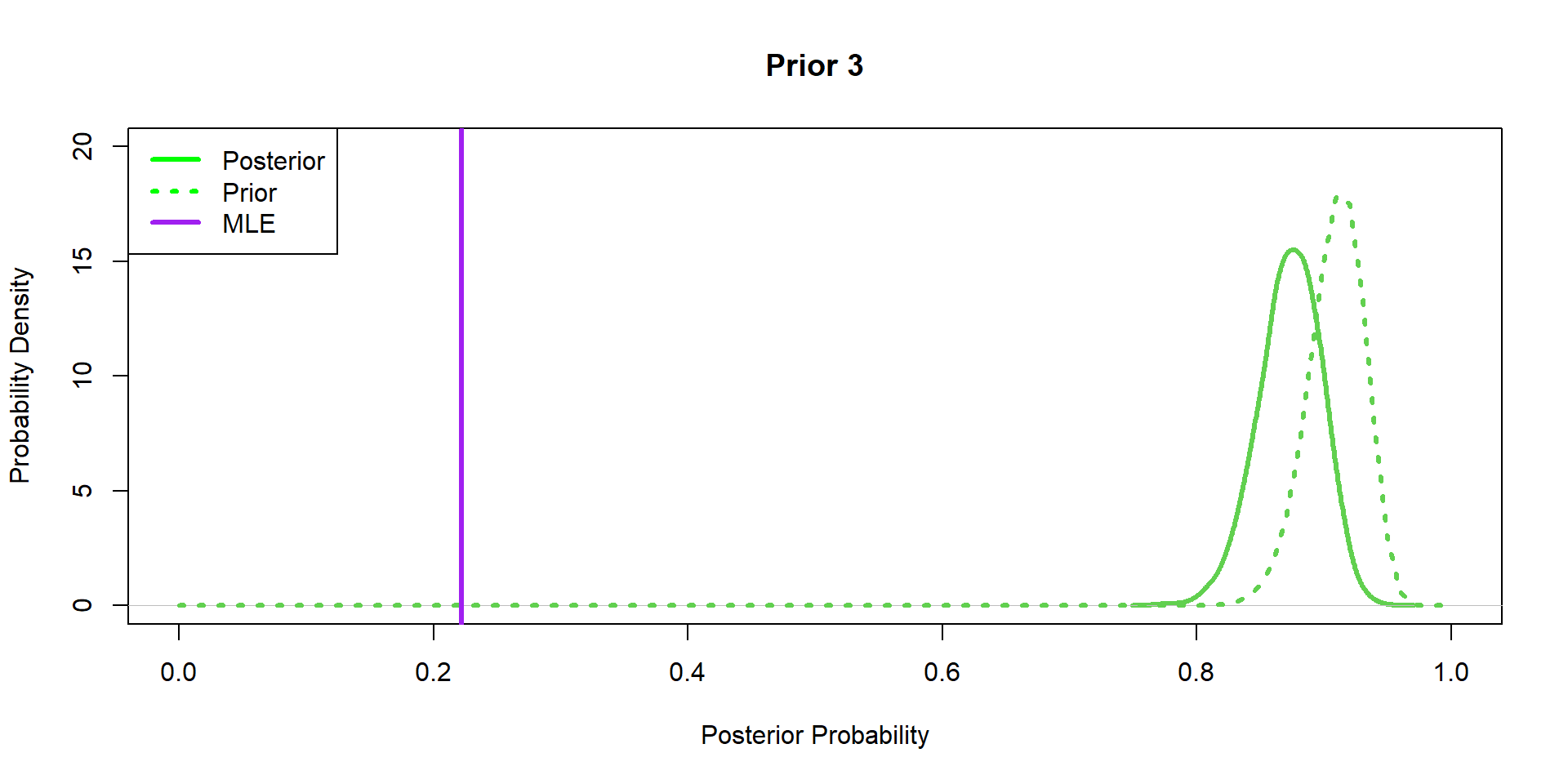
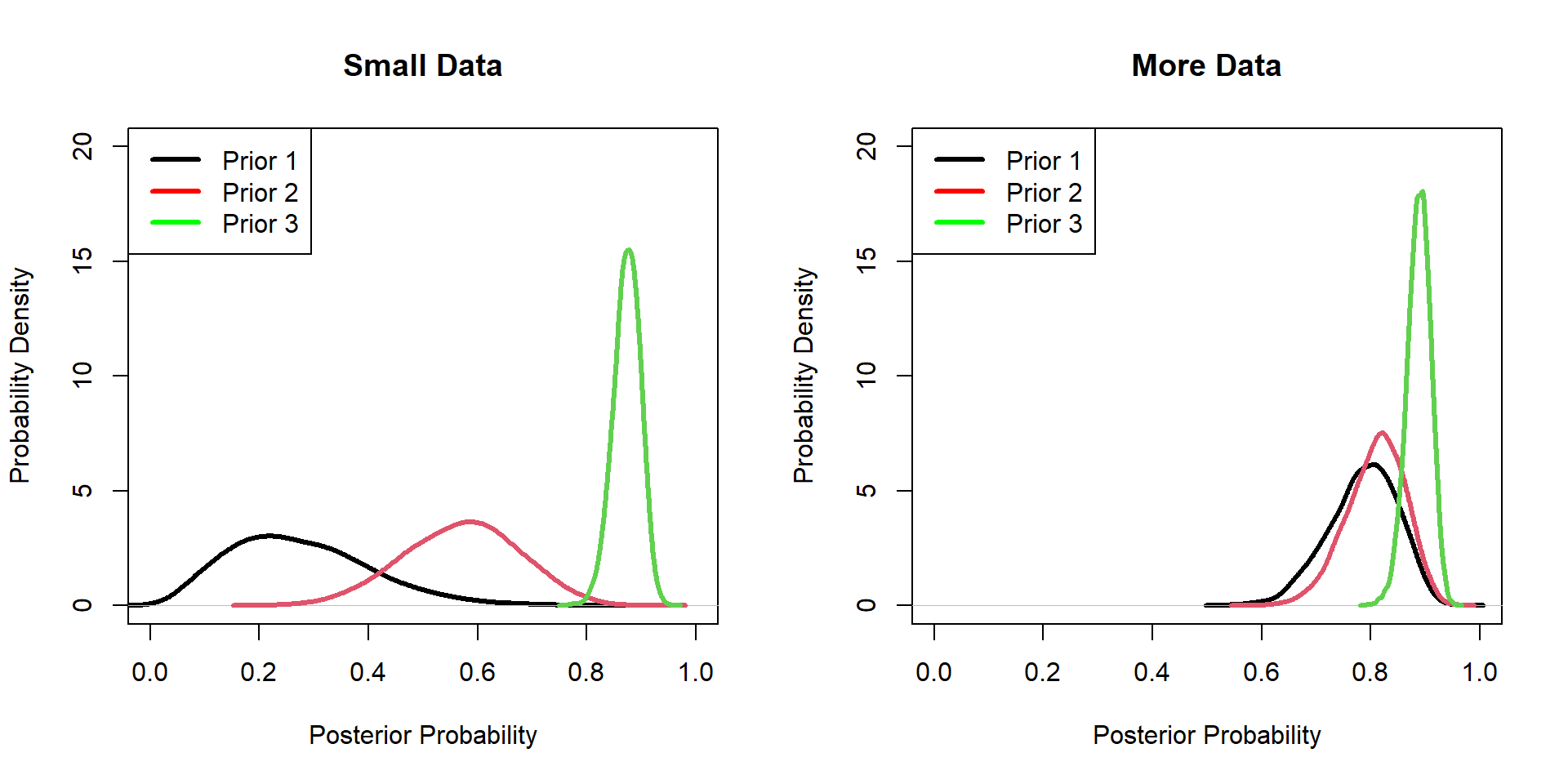
Hippos: More data! (Prior 1)
[1] 40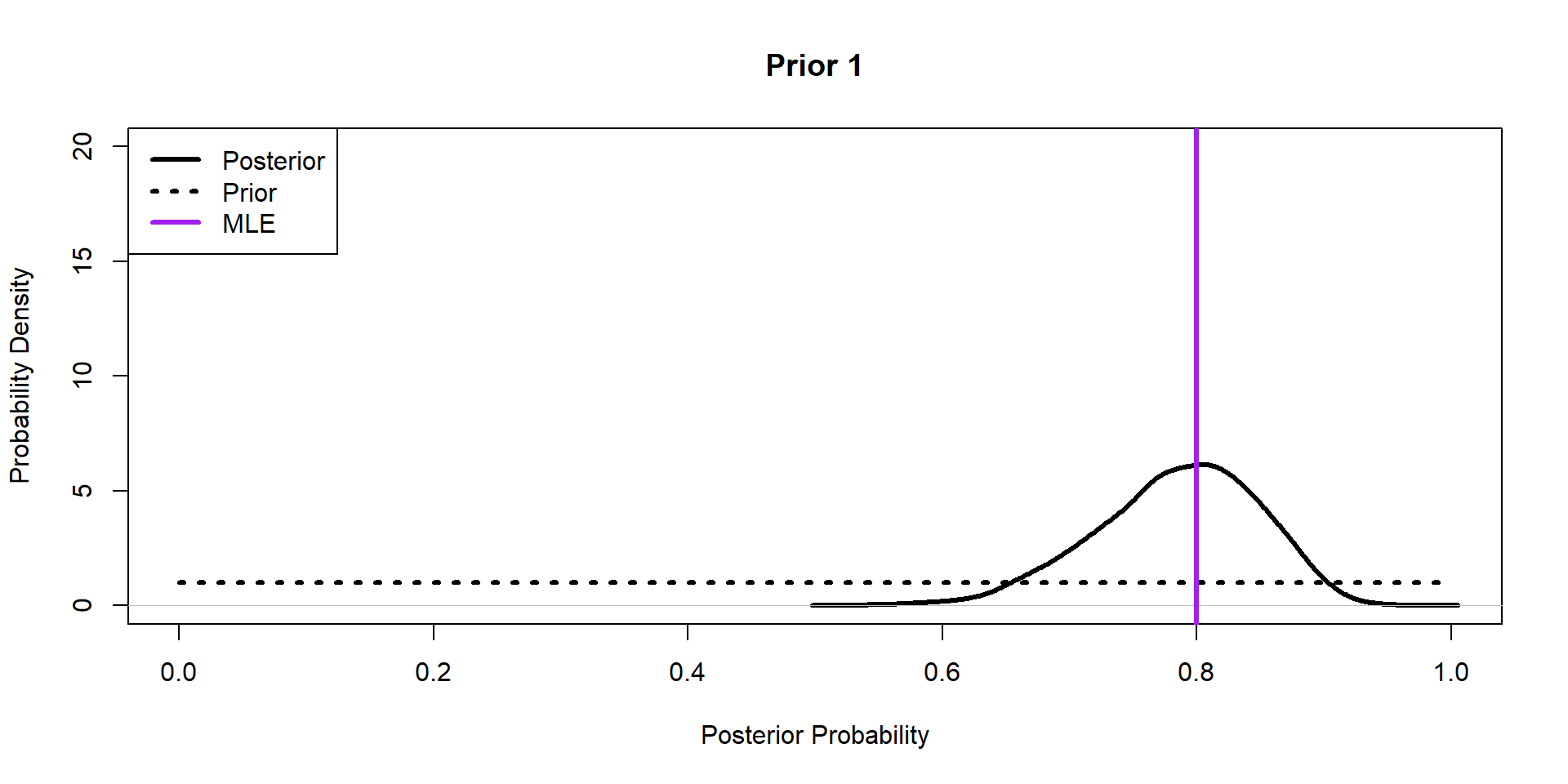
Hippos: More data! (Prior 2)
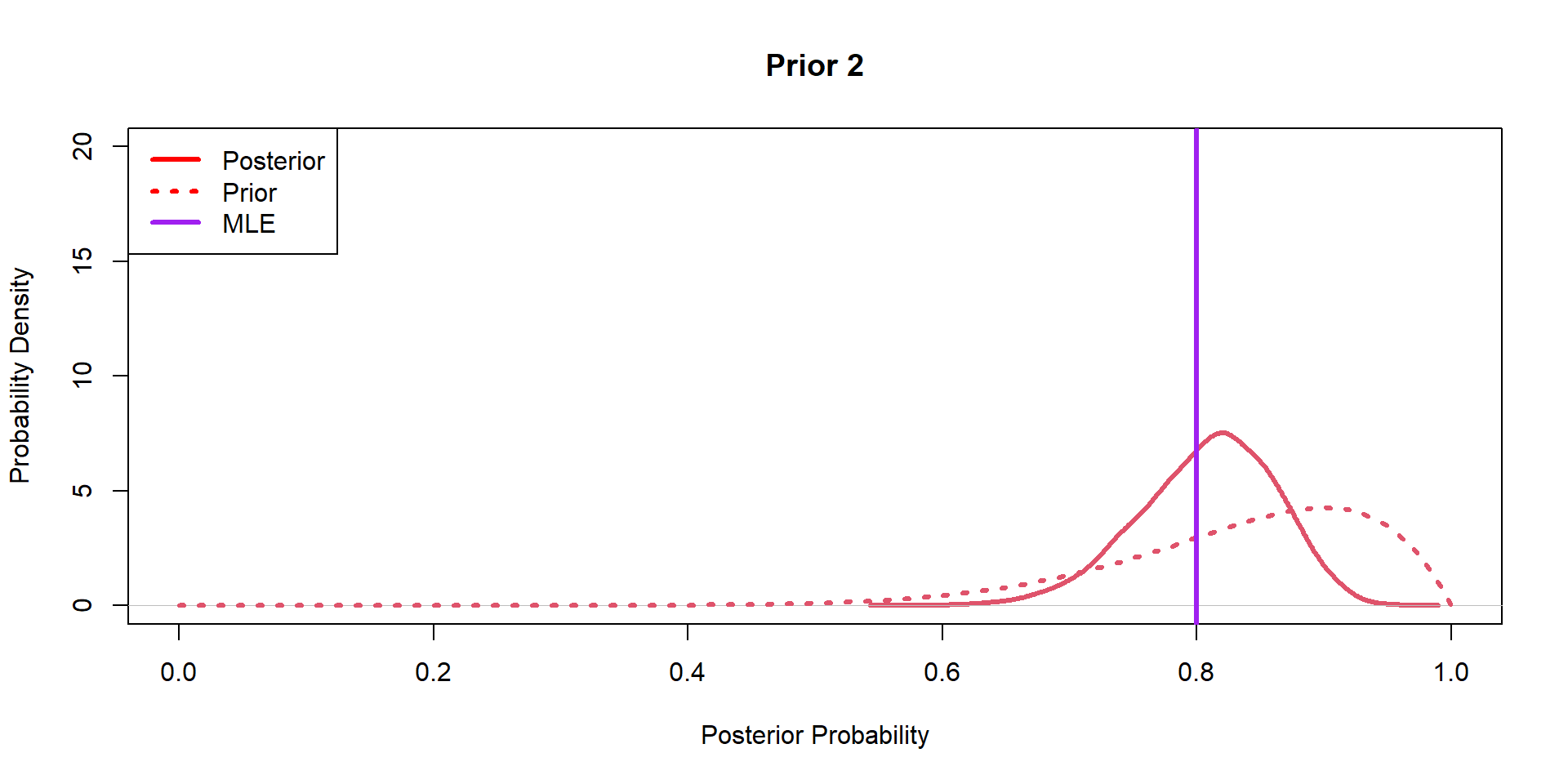
Hippos: More data! (Prior 3)
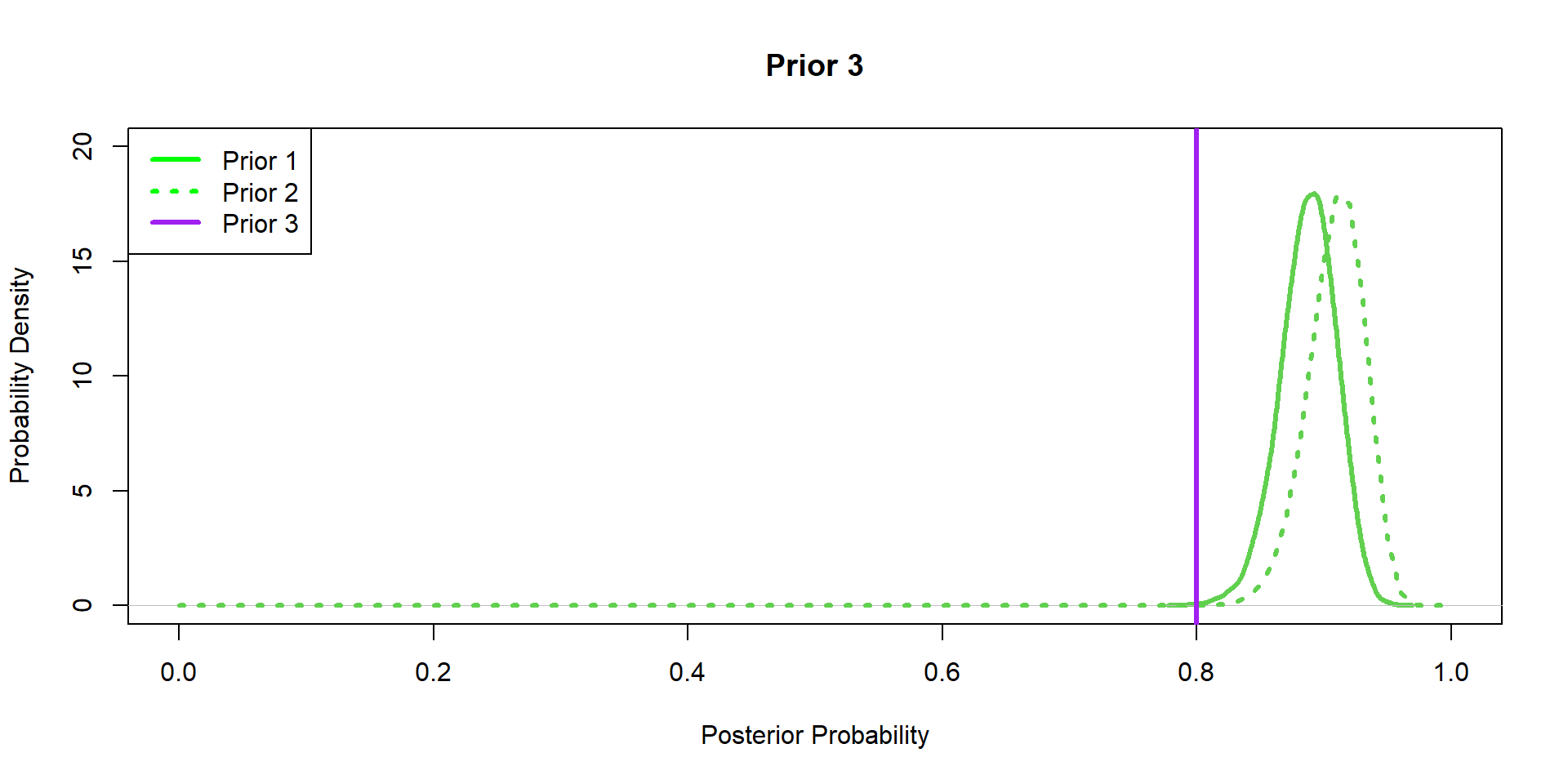
Hippos: Data/prior Comparison
Markov Chain Monte Carlo
Often don’t have conjugate likelihood and priors, so we use MCMC algorithims to sample posteriors.
Class of algorithim
- Metropolis-Hastings
- Gibbs
- Reversible-Juno
- No U-turn Sampling
\[ P(p|y) = \frac{\prod_{i=1}^{N}(p^{y}(1-p)^{1-y_{i}}) \times \frac{p^{\alpha-1}(1-p)^{\beta-1}}{B(\alpha,\beta)} }{\int_{p}(\text{numerator})} \]
\[ P(p|y) \propto\prod_{i=1}^{N}(p^{y}(1-p)^{1-y_{i}}) \times \frac{p^{\alpha-1}(1-p)^{\beta-1}}{B(\alpha,\beta)} \]
MCMC Sampling
- number of sample (iterations)
- thinning (which iterations to keep), every one (1), ever other one (2), every third one (3)
- burn-in (how many of the first samples to remove)
- chains (unique sets of samples; needed for convergence tests)
Sofware Options
- Write your own algorithim
- WinBUGS/OpenBUGS, JAGS, Stan, Nimble