[1] 3Wildlife Ecology Modeling
Class Stuff
Instructor: Brian Gerber
Classroom: NR 243
- Lecture/Lab/Discussion
When: Tu 11am - 12:15pm and Fr 10am - 1pm
My Office: 202A Wagar, Colorado Cooperative Research Unit
Office hours: TU 1:30pm - 2:30pm and by appointment
brian.gerber@colostate.edu
Computers: Bring laptop to class
- Software/R Packages should be installed prior to lab (posted on website)
Registration:
- 680A4 001 (CRN 77149) and 680A4 L01 (CRN 77150)
What is this course?
A mix of…
- statistics
- modeling
- coding
- math / notation
- science philosophy
- wildlife ecology and conservation
Assessment
| Assessment Components | Percentage of Grade |
|---|---|
| Course Engagement | 10% |
| Lab Assignments | 40% |
| Discussions | 10% |
| Quizzes | 10% |
| Project | 30% |
Project
independent or group research project - highlighting a modeling application, data/code transparency, and communication of results
group development of a lecture and lab case-study that showcases a statistical application relevant to wildlife ecology and conservation
Course Learning Objectives
Upon successful completion of this course students will be able to:
think ‘statistically’
read quantitative ecology literature
write code to fit and interpret complex statistical models relevant to wildlife ecology and conservation
communicate statistical approaches and results
Why is this class useful?
Able to read modern ecological literature
Understand what you are doing when using data and models; coding/statistics/modeling are related but not the same
Statistical modeling and coding skills are highly marketable
Taking control of your analyses
Collaborate with colleagues/statisticians
Software

Why learn to code?
- efficiency
- transparency
- flexibility in application
- shareable
- marketable skill
- needed for publications
Why use R?
- open-source and free
- small total user base / large in ecology and statistics
- find help online, e.g., stackoverflow
- data management
- statistics
- plotting / graphics
Why use RStudio?
- Makes using R easier
- Projects (file mgmt)
- R Shiny: Interactive online apps
- R Markdown: Interactive documents
- Quarto: interactive articles, websites, blog, …
- Posit - Certified B corp
Type of Modeling
parametric
probabilistic
generative
inferential and/or predictive
Statistics in the Modern Age
- Cox and Efron, Sci. Adv. 2017;3: e1700768.
Coding in the Modern Age
Software changes all the time
Code will become obsolete
Base R functions change slower than packages
Document/Annotate code and publish it online
File management is important; use sub-folders
A Graduate Student’s Dilemma
You need to know….
- field techniques
- logistics / planning
- people/advisor management
- institutional bureaucracy
- ecological theory
- wildlife mgmt principles
- conservation biology principles
- study design
- data management
- public speaking
- independent and team work
- graphic/visual arts
- ‘the literature’
- the job market
- how to write a manuscript/thesis
- writing/sharing coding
- statistical modeling
- …
Learning
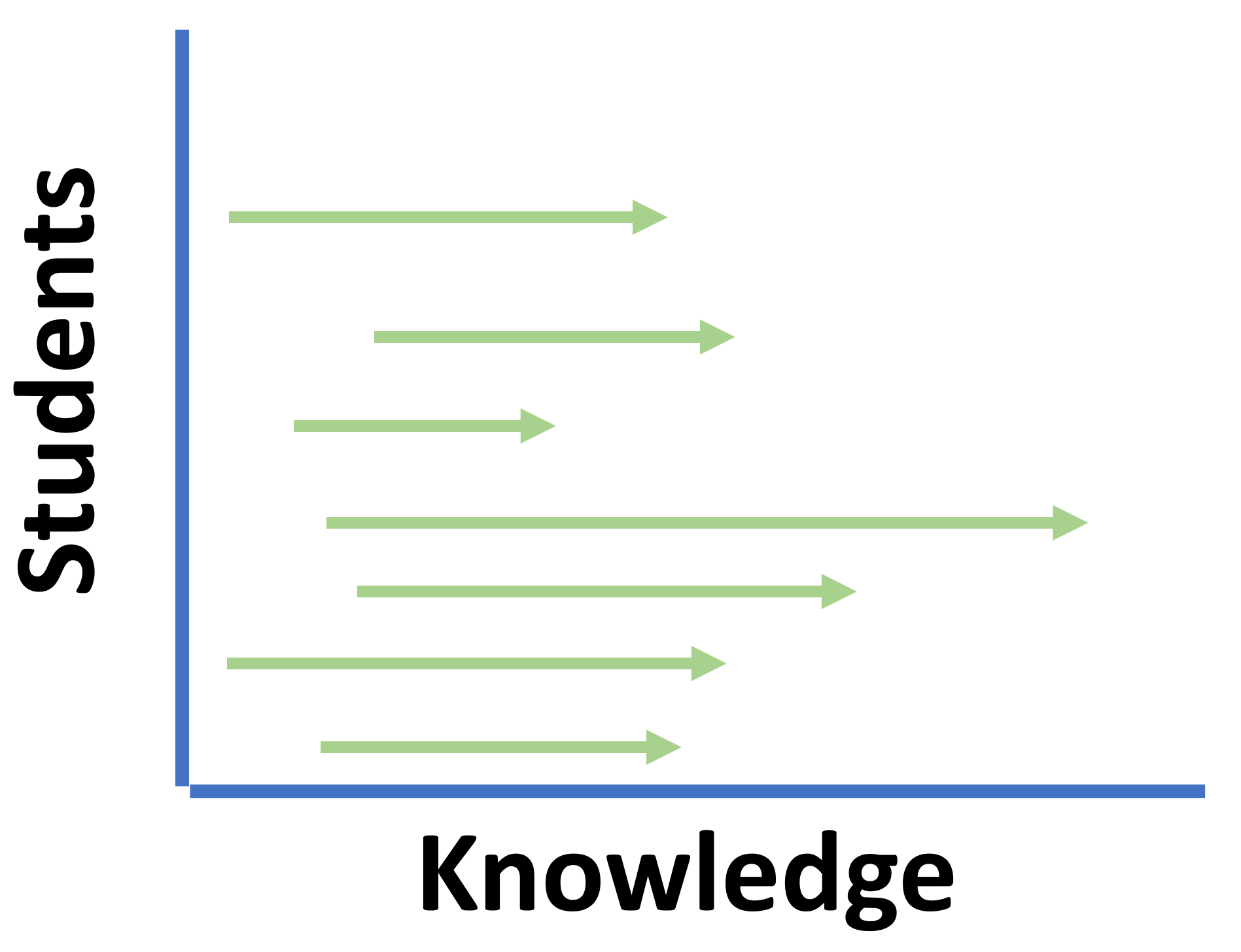
I do not know where you are starting
Our Aim
What model did you fit? Why? How?

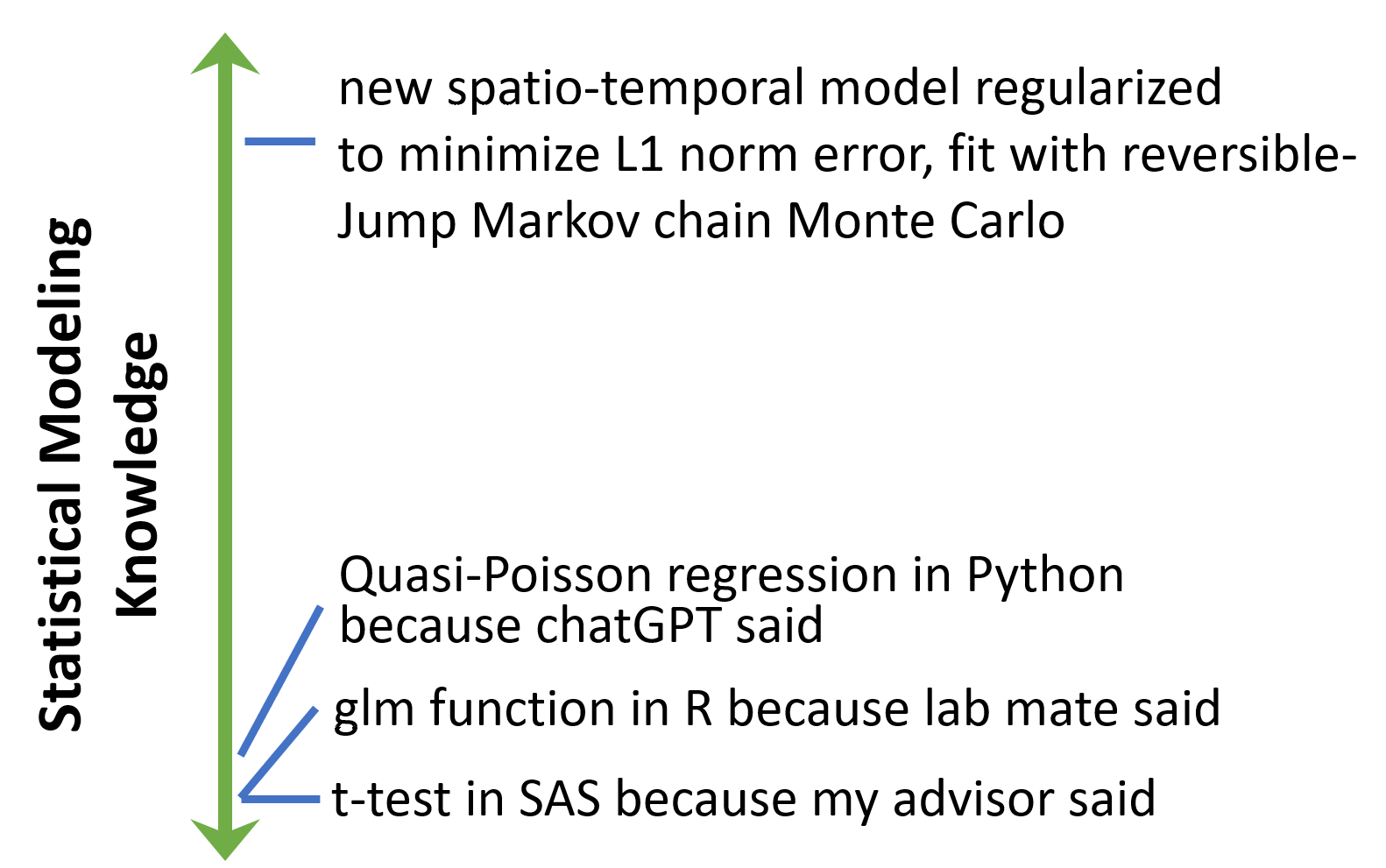
Think Statistically
Know…
your objective in fitting a model
the model and its properties (not just the name)
how to interpret ALL the parameters
how the parameters are being optimized
and have justification for modeling decisions
- requires reading literature
- requires evaluating assumptions yourself
Why is statistics and ecological modeling so difficult?
My Background

My Background

Science Practice
I am a pragmatist
There are many ways to do great science
There are more ways to do meh science
Disciplines have conventions
There are foundations of scientific and statistical learning
Know the why of your decisions
Ask lots of questions to everybody all the time
Teaching Philosophy
Learning is a choice (in every movement)
An inclusive environment is paramount for learning
Communication is key
Everyone has something to teach and something to learn
Struggle is good. Solving problems leads to learning
BUT….
Website
Class Questions

RStudio
What does each panel do?
The language of R
Objects
A storage place for information; stored in the “Environment”
‘Attributes’ describes the structure or information of the object
The language of R
Objects
The language of R
Values
- numeric
- integer
- character
- factor
Objects
- vector
- matrix
- array
- list
- dataframe
- S3, S4, S5, and beyond
The language of R
Functions
‘does stuff’; creates or manipulates objects
‘Arguments’ are the types of things a function is asking for; the inputs
The language of R
object = function(argument1 = input1, argument1 = input2)
object = function(input1, input2)
Some useful functions
for loops
Some useful functions
Create your own function
Some useful functions
apply/sapply/lapply/vapply
[1] -0.356772536 0.033934457 -0.237817472 -0.272299554 -0.494920197
[6] -0.049415000 -0.021611812 -0.005851695 -0.412304818 0.234196891Code Organization
Hierarchical code organization
- code structure using indenting
- top –> bottom execution
Help! My code doesn’t work…
cor.sp.route.cor=vector("list",n.species)
cor.sp=rep(NA,n.species)
for(s in 1:n.species){
route=new.cov.species.long.scaled[[s]]$routeID
cor.sp[s]=cor(patch.size20.species.scaled.mat.center.route[s,],patch.count20.species.scaled.mat.center.route[s,])
for(i in 1:nroutes){
temp1=patch.size20.species.scaled.mat.center.route[s,which(route==route.id[i])]
temp2=patch.count20.species.scaled.mat.center.route[,][s,which(route==route.id[i])]
if(length(temp1)>5){
cor.sp.route.cor[[s]]=abs(c(cor.sp.route.cor[[s]],cor(temp1,temp2)))
}}}Better…
# Create Storage objects
cor.sp.route.cor=vector("list",n.species)
cor.sp=rep(NA,n.species)
#loop over species
for(s in 1:n.species)
{
route=new.cov.species.long.scaled[[s]]$routeID
cor.sp[s] = cor(patch.size20.species.scaled.mat.center.route[s,],
patch.count20.species.scaled.mat.center.route[s,]
)
# loop over species and routes
for(i in 1:nroutes)
{
temp1 = patch.size20.species.scaled.mat.center.route[s,which(route==route.id[i])]
temp2 = patch.count20.species.scaled.mat.center.route[,][s,which(route==route.id[i])]
if(length(temp1)>5){
cor.sp.route.cor[[s]]=abs(c(cor.sp.route.cor[[s]],cor(temp1,temp2)))
} #End if statement
} #End routes loop
} #End species loop